The present Solution aims to establish environmental DNA and RNA (eDNA and eRNA), coupled with advanced DNA sequencing technology (metabarcoding), as a new tool to evaluate the presence of pathogens in aquatic facilities. This technology has the potential to reduce the mortality and morbidity of animals. Collaborators are sought in any sector using aquatic animal health monitoring to assess and validate this eDNA metabarcoding technology against current practice. Input from partners who could develop and validate software, to contain searchable DNA sequences of pathogens, are also welcomed.
Aquatic models (cephalopods, fish and amphibians) are used in diverse biomedical contexts, and numerous efforts are made to define guidelines for the husbandry, housing, welfare and health status of these animals (Felasa et al., 2015). Improving animal health and welfare raises the quality, consistency and reliability of experiments involving animals, enabling better interpretation of experimental results.
The presence of pathogens is routinely used as a measure of animal health in aquatic facilities. Panels of species-specific pathogens and protocols for their identification are available for assessing the health of some species, i.e. zebrafish (Collymore et al., 2016), Xenopus (Alworth et al., 2007), and to a lesser degree, Cephalopods (Fiorito et al., 2015). These approaches often use pre- and post- filtration sentinels, moribund fish and symptomatic animal groups, and require samples to be taken from the animals (e.g. necropsy and histopathology of tissues and organs). The number of sentinels used varies according to the facility size, but recent evidence suggests at least six sentinels to be screened every three to six months (Mocho et al., 2017).
Recently real-time PCR, conventional PCR and matrix-assisted laser desorption–ionization time-of-flight mass spectrometry (MALDI-TOF MS) have been adopted to identify aquatic pathogens in sludge samples from the animal’s environment (Mocho 2016). These approaches require a threshold of contamination to be reached before the pathogens are detected, so they are complementary tools to the routine histopathological sentinel screening used to monitor the health of the colony (Collymore et al., 2016; ZIRC 2009; Mocho et al., 2017).
References:
- Alworth LC, Harvey SB (2007). IACUC Issues Associated with Amphibian Research. ILAR Journal 48(3): 278–289. doi: 10.1093/ilar.48.3.278.
- Collymore C, Crim MJ, Lieggi C (2016). Recommendations for Health Monitoring and Reporting for Zebrafish Research Facilities. Zebrafish 13 Suppl 1: S138-48. doi: 10.1089/zeb.2015.1210.
- FELASA working group. Available at: http://www.felasa.eu/working-groups/working-groups-present/zebrafish-housing-husbandry-and-health-monitoring-recommendations/.
- Fiorito G, Affuso A, Basil J, et al. (2015). Guidelines for the Care and Welfare of Cephalopods in Research -A consensus based on an initiative by CephRes, FELASA and the Boyd Group. Lab Anim 49(2 Suppl): 1-90. doi: 10.1177/0023677215580006.
- Mocho JP, Martin DJ, Millington ME, et al. (2017). Environmental Screening of Aeromonas hydrophila, Mycobacterium spp., and Pseudocapillaria tomentosa in Zebrafish Systems. J Vis Exp (130): 55306. doi: 10.3791/55306.
- Mocho JP (2016). Three-Dimensional Screen: A Comprehensive Approach to the Health Monitoring of Zebrafish. Zebrafish 13 Suppl 1: S132-7. doi: 10.1089/zeb.2015.1200.
- ZIRC, Zebrafish International Resource Center (2009). Sentinel fish program. Available at: https://zebrafish.org/documents/protocols/pdf/health_monitoring/sentinel_fish_program. (Accessed 7 Mar. 2018).
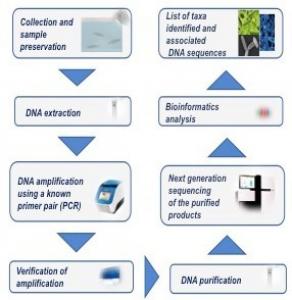
Researchers at the University of Naples Federico II have developed a non-invasive, cost efficient method to detect and identify potential pathogens (viruses, bacteria, protozoa, metazoa, parasites) in aquatic facilities. eDNA can be extracted from water samples collected (few millilitres) using a sterile pipette from all pre-filtration tanks and three post-filtration tanks on each rack (in a recirculating system) during routine facility monitoring. The eDNA is amplified by PCR using primers either specific for a target species or generic for multiple-taxon analysis and then sequenced (Thomsen et al., 2015). The obtained sequences are compared to the GenBank international reference database using bioinformatics tools. This process is known as metabarcoding and enables the rapid identification of DNA barcodes (short DNA fragments specific for a species) from the total DNA extracted from the environmental sample. A gap remains between detection of a microbe in the environment and the impact of the potential pathogen on animals, but increased use of this technology will lead to an increased understanding of the links between a microbe's prevalence and an animal's disease.
Aquatic organisms (such as Cephalopods, fish and amphibians) are sensitive to a wide range of cues (handling, vibration, chemicals, etc) so this Solution could be used to refine routine health monitoring by:
- Ensuring a bimonthly non-invasive evaluation of the microbiological status of aquatic organisms.
- Identifying opportunistic infection and emerging pathogens.
- Standardising the health monitoring program and reporting format in multispecies aquatic facilities, specifically those containing species emerging as model organisms for study.
All the above-mentioned actions enable improved identification of pathogens in local and imported animals, supporting quarantine and bio-security measures, preventing potential pathogens spreading to healthy animals, leading to enhanced animal welfare.
eDNA metabarcoding could be coupled with software containing DNA/RNA sequences of pathogen species. This searchable database would enable users to easily identify the potential pathogens by filtering the pathogen barcode sequences according to the animal species maintained in the facility, reducing the time taken for pathogen identification.
eDNA metabarcoding can also be applied to the litter and faeces of terrestrial animals, for the analysis of many habitats (water or soil), food sources, and identification of new animal organisms (Bruno et al., 2015).
Figure 1. Workflow of the eDNA metabarcoding
References:
- Bruno, F, Marinella M, Santamaria M (2015). e-DNA meta-barcoding: from NGS raw data to taxonomic profiling. Methods Mol Biol 1269: 257-78. doi: 10.1007/978-1-4939-2291-8_16.
- Thomsen PF, Willerslev E (2015). Environmental DNA – An emerging tool in conservation for monitoring past and present biodiversity. Biological Conservation 183: 4-18. doi: 10.1016/j.biocon.2014.11.019.
Partners in any sector using aquatic animal health monitoring are sought to validate this technology. After validation of the eDNA metabarcoding technology, the collaborators can use the device to routinely screen the microbiological status of their aquatic organisms, with free access to the associated software.
Potential partners should be familiar with current methodologies (e.g. PCR and microbial MALDI-TOF MS) and willing to work with researchers from the University of Naples Federico II to assess and validate this eDNA metabarcoding technology against their current practice. Collaborators would be required to send either water samples collected in their facilities or amplified and sequenced DNA/ RNA fragments to blast in the software, enabling identification of the pathogen species. Networks of multispecies aquatic facilities able to provide samples and access to facilities would also be valuable for validation studies.
Input from partners who could develop and validate software to contain searchable DNA sequences of pathogens would also be welcomed.
The presence of pathogens is particularly important when importing/ exporting animals, as it prevents the transport of animals with subclinical infections that could become lethal upon stress induced by transport. This technology can also prevent infection spreading to the new facility, by avoiding the movement of morbid animals or by raising quarantine and bio-security measures. This refinement has the potential to lead to a reduction in the number of sentinels required, because the increased monitoring and reaction to the presence of pathogens in the water leads to reduced mortality and morbidity in the colony. In the technology developer’s multispecies aquatic facility this technology has reduced the number of sentinels required by approximately 50%. For example, within their zebrafish facility application of this technology has reduced the number of sentinels required from 12 to six animals. Scaled up across aquatics facilities globally this would represent a substantial reduction in animal use. For example, in the UK alone there are around 100 aquatic facilities and if they each use a minimum of 12 sentinels, there is the potential to reduce the number of sentinel animals required across the UK by 600 for each health assessment screening.
Additionally, through increased use of this technology links between a microbe's prevalence and an animal's disease are hoped to be established, which has the potential to reduce (and hopefully replace) the need for sentinels to understand the health status of the colony in the future.