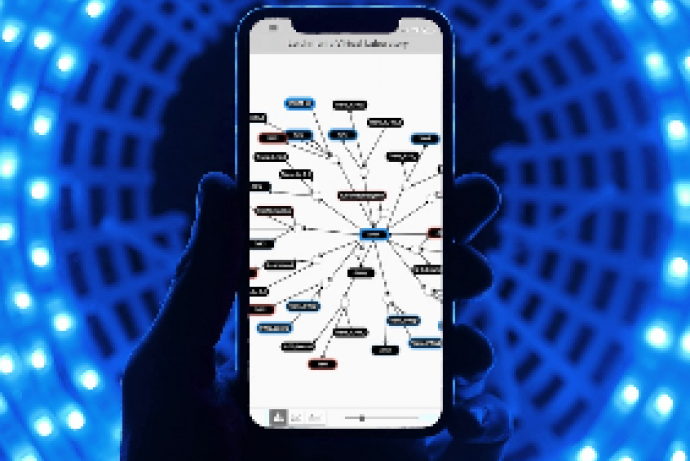
Insights from computational and mathematical (in silico) models can reduce the number of animals used in the development of new products and drug treatments. To accelerate the adoption of in silico models and maximise their 3Rs impact, confidence and transparency in model outputs is key to provide scientific insights and support decision making. Simomics’ Virtual Disease Laboratory technology enables effective exploitation of in silico models, providing an online platform to explore, evidence, share and communicate the model and its results. Simomics is seeking collaborators with in silico disease models with 3Rs benefits to be enhanced through Virtual Disease Laboratories.
Employing in silico models is one strategy for reducing the number of animals used in the development of new therapeutics, combination therapies, biomarkers and medical devices. The models can be used to gain insights into many stages of the development process: understanding disease progression and dynamics; identifying novel therapeutic targets; investigating delivery dynamics and therapeutic combination strategies; and predicting unwanted side effects and toxicities.
However, applying in silico models to inform development of new products and drug treatments faces many challenges. Models can often be impenetrable to non-modellers and scientists from other disciplines. Better communication of how a model works and its scope (the conditions under which its outputs are applicable) should lead to better decisions about how model outputs can inform the science and any 3Rs benefits that result. The ability to clearly communicate a model and its outputs is key both within an organisation between colleagues, and externally at the regulatory interface.
Developing good quality in silico models requires a great deal of effort and skill. Often models are developed by individual modellers and when they leave their position, important details can be lost to the organisation. These include how and why the model was designed the way it was (assumptions and abstractions), justifications for parameter settings, and the scope of the model. This makes it difficult to revisit the model in future, and to reuse or repurpose the model for a different scenario.
A wide range of tools and techniques are typically required to effectively develop, use and maintain in silico models. The models come in many formats (e.g. equation-based or agent-based) and may be implemented in different programming languages and environments. Models must be parameterised using suitable values, which need to be extracted from a range of sources including scientific literature, in vivo or in vitro studies, or expert opinion. Statistical measures are required to calibrate models and analyse results. Capturing and communicating these design decisions so that they can be audited would help to ensure model outputs are trusted.
Simomics develops tools to support evidence-based decision making from in silico models with a focus on reducing the number of animals used in the development of new products and drug treatments. Here a unique solution is offered for improving the reuse and communicability of in silico models: Virtual Disease Laboratories, which are custom applications integrating Simomics evidencing tools with client data and in silico models.
The development of in silico model code requires a range of design decisions to be made, many of which go unrecorded. Such design decisions include choice of modelling tools and techniques, abstractions and assumptions from the modelled biological system, and choice of suitable model parameters. Virtual Disease Laboratory technology allows you to capture these details as evidence statements and then directly link where it is applicable in the model source code. Evidence statements can link to external supporting resources such as research publications, web links, images, or data files. Summaries of evidence can be visualised to assess evidence quality across code files, reveal evidence dependencies, and provide summary evidence statistics detailing quality of evidence and sources.
Virtual Disease Laboratories are customised by Simomics to provide appropriate visualisations of the in silico model structure and results from in silico experiments. Examples include graphical representation of model structure and how model behaviours change from varying model parameters. This allows the user to explore the model and its results in an intuitive way without having to analyse model code. As well as all types of application (target identification, toxicity, disease progression modelling) this system can integrate all types of data including e.g. genomic, proteomic, and chemical data as well as pathway and network analysis.
The Virtual Disease Laboratories are implemented as password-protected web accessible tools. This provides an open and transparent platform with model code evidencing and results visualisation in one place, which can be shared with colleagues and collaborators. Together this facilitates:
- The exploration of the model and results;
- Confidence of model outputs from transparent evidencing;
- Curation, auditing and validation of that model;
- Increased productivity through clear communication and retention of model IP.
Simomics has developed 3Rs-based Virtual Disease Laboratories to investigate disease dynamics for Leishmaniasis (https://www.leishsim.org) and Sarcoidosis, in addition to a Virtual Disease Laboratory for investigating fish ecotoxicology (https://vfetl-demo.simomics.com/guest).
A Virtual Disease Laboratory will enhance and improve how an in silico model can be used to inform decision making in an organisation. Simomics seek a collaborator(s) with an existing or in-development in silico model that is used within drug/product development and provides a 3Rs impact. The collaborator will need to have access to an in silico model and any associated code files. Preferably the model will be representable in Systems Biology Markup Language (SBML), but Simomics is open to collaborators with models written in most languages. Simomics will work with the collaborating model developer to evidence and visualise the collaborators model and will require an element of their time to build the Virtual Disease Laboratory to suit their needs. Access to model output results data would also be beneficial.
The collaborator will become an early adopter and be at the forefront of using this exciting 3Rs technology. At the end of the collaboration they will be in possession of a well evidenced model and associated visualisations that is embedded within a web-deployed tool. This can be used within their organisation where appropriate to aid evidenced decision making.
Information about IP
Simomics current IP covers techniques to evidence code, analysis of evidence coverage, transformation of code to graphical representations, Virtual Disease Laboratory user interface elements, and Virtual Disease Laboratory tool infrastructure. Any in silico model code and evidence/results data integrated into a Virtual Disease Laboratory will remain with the collaborator.
Virtual Disease Laboratories facilitate and maximise the 3Rs impacts achievable with in silico models. Well-constructed, evidenced and transparent models can replace and reduce animal experimentation. Replacement benefits arise from in silico models being used to understand the science and data behind disease, drug interactions, dosing and toxicity instead of using animal experiments (Kovatchev, et al. 2009). Reduction benefits arise from better experimental design, whereby in silico models and data analyses are used to minimise the number of animals per experiment, or to narrow the range of experiments that need to be performed (Jean-Quartier, et al. 2018).
Quantification of 3Rs impacts from in silico approaches varies per project. With LeishSim (https://www.leishsim.org), a model was used to identify potential Leishmaniasis parasite drug targets by performing a series of knock-out experiments on the range of molecular species (receptors, cytokines etc.,) in the model. Of the 96 targets, only one was deemed promising enough to justify in vivo validation. Each target ruled out for in vivo validation saved between 10 and 20 animals. The Virtual Disease Laboratory infrastructure used in LeishSim improved model quality and confidence in the results by facilitating model analysis and communication between modeller and wet-lab colleagues.
References
Kovatchev B, et al. (2009). In Silico Preclinical Trials: A Proof of Concept in Closed-Loop Control of Type 1 Diabetes. Journal of Diabetes Science and technology; doi.org/10.1186/s12885-018-4302-0.
Jean-Quartier C, et al. (2018). In silico cancer research towards 3R, BMC Cancer (1), doi:10.1177/193229680900300106.