The Neuroinformatics lab at Newcastle University has developed Vertex, a computational model of mammalian brain tissue that can reproduce patterns of recordings observed in ex vivo tissue. Using Vertex it is possible to study the effect of stimulation on the activity in different cortical layers. Stimulation can be electric, magnetic, optogenetic, or through ultrasound. Pharmacological intervention, leading to changes in ion channel properties, can also be simulated in the modelling framework. This model, validated through comparisons with experimental recordings, may replace some animal use during the early stages of medical device development or pharmacological drug discovery. Industrial and academic collaborators are sought with expertise in various brain diseases and/or safety and efficacy of drug development to explore and expand the capabilities of the model.
There is increasing recognition globally of the potential impact computational models could have on drug discovery and biomedical device development (Viceconti et al., 2016; Kovatchev et al., 2009). Current models of neural systems range from large-scale simulations of a billion neurons, such as the SpiNNaker system that is linked to the Human Brain Project, to more detailed models of single neurons or small populations of neurons. However, these models either do not generate the type of signals that can be measured experimentally (local field potentials) and instead focus on reproducing the activity patterns (spiking behaviour) of neurons, or they do not represent the anatomical and physiological properties of ex vivo or in vitro brain tissue models.
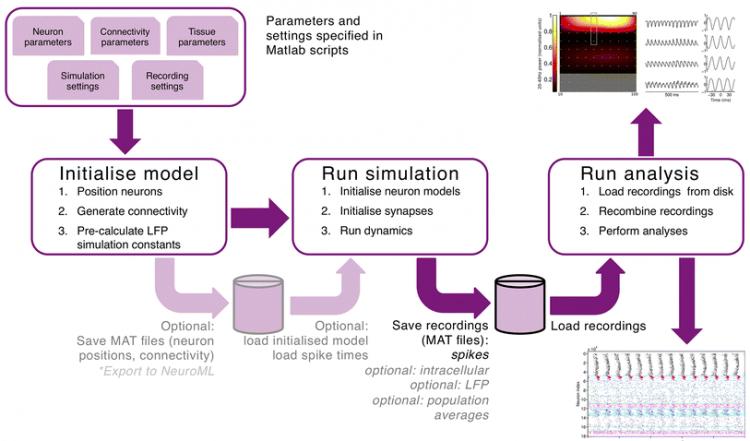
Developing computer models of brain tissue, based on literature and online information of the connectivity and morphology of neurons and the layer architecture of brain regions, is a major endeavour that previously was focused on rodent (mouse) brains and detailed modelling of individual neurons and small circuits. The Vertex simulator has been developed to contain fewer details at the level of individual neurons, but allows for a rapid simulation of activity at the cortical tissue level in species including rat, ferret and rhesus monkeys encompassing 100,000s of neurons. In addition, the Vertex software allows the user to measure the local field potential, as recorded by electrodes placed within the tissue, at any point within the three-dimensional tissue simulation. In this way, the recording from the Vertex computer model can be directly compared to an experimental recording in ex vivo brain tissue. Moreover, the model can be extended to predict in vivo recordings from the top of the cortical surface (electrocorticogram) or non-invasive electroencephalogram recordings (EEG).
References:
- Kovatchev BP, Breton M, Man CD, et al. (2009). In silico preclinical trials: a proof of concept in closed-loop control of type 1 diabetes. Journal of Diabetes Science and Technology 3(1): 44-55. doi: 10.1177/193229680900300106.
- Viceconti M, Henney A & Morley-Fletcher E (2016). In silico Clinical Trials: How Computer Simulation will Transform the Biomedical Industry. Research and Technological Development Roadmap. Brussels: Avicenna Consortium. Avicenna Consortium, Brussels 3(2): 37-46. doi: 10.18203/2349-3259.ijct20161408.
Collaborators who have expertise in various brain diseases, and different animal species and brain tissues are being sought to test the reach of the model. Alliances are also being sought with pharmaceutical and biotechnology companies to explore the utility of the computational models in improving safety and efficiency of drug development aimed at brain disorders.
Partnerships will help us to identify areas with unmet needs and how the specification of this Solution needs to be adapted to fulfil them. The Solution providers will provide their expertise and software to any commercial institution, pharmaceutical company, or medical device company to conduct exploratory experiments or test the functional effects of their interventions using the computational platform.
In Great Britain in 2016, over 200,000 experimental procedures were carried out on animals, including mice, rats, ferrets and primates (all species that can be modelled using Vertex) to study the nervous system (Home Office, 2017). It is difficult to accurately determine how many of these animals were used in studies that could have adopted the Vertex model, but it is clear from literature searches that ex vivo brain slice recordings play a significant role in neurobiology. In some studies, as many as 34 animals have been used to obtain brain tissue (Rohan et al., 2015). Using Vertex instead, as a predictive tool to pre-screen potential compounds, could play a significant role in reducing some of this animal use and improving the translation of drugs into the clinic.
Fewer than 10% of drugs that show potential in animal studies and progress towards clinical trials ever get licensed for human use (Manolis et al., 2013); a proportion that is even lower for brain diseases. If only half of the compounds that later fail could be identified earlier using the Vertex simulator and removed before vertebrate testing, the number of experiments requiring animals and/ or primary tissue would be significantly reduced.
References:
- Home Office (2017). Annual Statistics of Scientific Procedures on Living Animals, Great Britain 2016.
- Manolis E, Rohou S, Hemmings R, et al. (2013). The Role of Modeling and Simulation in Development and Registration of Medicinal Products: Output From the EFPIA/EMA Modeling and Simulation Workshop. CPT: Pharmacometrics and Systems Pharmacology 2(2): e31. doi: 10.1038/psp.2013.7.
- Rohan J, Carhuatanta K, McInturf S, et al. (2015). Modulating Hippocampal Plasticity with In Vivo Brain Stimulation. Journal of Neuroscience 35(37): 12824-12832. doi: 10.1523/JNEUROSCI.2376-15.2015.
Vertex provides a novel simulation platform and research tool for estimating the effect of intervention (electrical, magnetic, ultrasound, drugs) on brain tissue by mimicking the behaviour of individual neuron types (spike trains) or effects at the population level (local field potential). The model covers both immediate effects and longer-term effects caused by synaptic plasticity. The model can be adapted to different species (rat, mouse, human) or different types of brain tissue (e.g. hippocampus, retina, cortex). However, due to limited information on connectivity, the human model is currently more restricted. Species specific versions of the Vertex tissue model have been demonstrated to reproduce the ex vivo behavior of both rhesus monkey cortical tissue (Tomsett et al., 2015) and the effect of electrical stimulation of ex vivo ferret brain tissue (Frohlich & McCormick, 2010). Applying the model to drug discovery may lead to fewer failures late in the drug development pipeline as effects on brain tissue can be tested in a system based on the primate brain (rhesus monkey), closer to humans than the current rodent animal models (Vallender & Miller, 2013). Indeed, CNS drugs show the highest failure rate at the human clinical phase (34%) compared to other organs, while the failure rate of 7% at the preclinical animal phase is lower than for other organs (Cook et al., 2014). It is also possible to easily include gene regulatory circuits within the model if this level of detail is needed.
Figure 1. Parameters and workflow of a brain tissue simulation leading to recordings of the local field potential (top, showing power in the gamma range) across layers and of spiking neuronal activity across time (bottom).
Vertex opens the possibility to test which interventions have a maximal effect while reducing unwanted side effects in a brain model for multiple species (rat, mouse, ferret, primate and human). In this way, only the most promising interventions would progress to regulatory required animal studies; reducing the costs of drug development for pharmaceutical companies and the number of early-stage animal experiments. Furthermore, computational models might indicate the most suitable species for later animal experiments.
The system could also be used for medical device companies to test the effect of brain stimulation devices, e.g. implantable devices using optogenetics or electricity, before starting trials with animal models. In this way, both costs and time to market would be reduced. The researchers expect that computer simulations concerning the safety of devices will become part of the regulatory process and therefore this platform could be one component of complying with regulatory authorities in the UK and abroad.
References:
- Cook D, Brown D, Alexander R, et al. (2014). Lessons learned from the fate of AstraZeneca's drug pipeline: a five-dimensional framework. Nat Rev Drug Discov 13: 419-431. doi: 10.1038/nrd4309.
- Frohlich F. & Mccormick DA (2010). Endogenous electric fields may guide neocortical network activity. Neuron 67(1): 129-43. doi: 10.1016/j.neuron.2010.06.005.
- Tomsett RJ, Ainsworth M, Thiele A, et al. (2015). Virtual Electrode Recording Tool for EXtracellular potentials (VERTEX): comparing multi-electrode recordings from simulated and biological mammalian cortical tissue. Brain Struct. Funct 220(4): 2333-2353. doi: 10.1007/s00429-014-0793-x.
- Vallender EJ, & Miller GM (2013). Nonhuman primate models in the genomic era: a paradigm shift. ILAR J 54: 154-165. doi: 10.1093/ilar/ilt044.