CleanCut
The aim of this Challenge was to develop an in vitro model to replace in vivo tumourigenicity studies for safety assessment of genome edited human haematopoietic stem cells (hHSCs).
A team led by Dr Alessandro Rotilio from MOAB srla have developed a dual chamber bone marrow and lymph node bioreactor platform for assessing the tumorigenicity of gene-edited human haemopoietic stem cells (hHSCs) through the CleanCut Challenge.
Publication
Ritter, P. et al. (2024) ‘A millifluidic bioreactor allows the long term culture of primary lymphocytes or CD34+ hematopoietic cells while allowing the detection of tumorigenic expansion’, Frontiers in Bioengineering and Biotechnology, 12. doi:10.3389/fbioe.2024.1388312.
Challenge Completed
A team led by Dr Alessandro Rotilio from MOAB srla have developed a dual chamber bone marrow and lymph node bioreactor platform for assessing the tumorigenicity of gene-edited human haemopoietic stem cells (hHSCs) through the CleanCut Challenge.
Phase 2 awarded
A team led by Dr Alessandro Rotilio, MOAB srla, a spin-off of Politecnico di Milano incubated by Hub-Initio, has been awarded £1,000,000 to deliver Phase 2.
Read more about MOAB on their website.
Phase 1 Winners
Project teams led by:
- Dr Uwe Marx, TissUse GmbH, Germany, £99,960.
- Professor Manuel Salmeron-Sanchez, University of Glasgow, £99,278.
- Professor Manuela Raimondi, Politecnico di Milano, Italy, £98,824.
Challenge launched
Sponsored by Novartis, Bayer and Takeda, this Challenge aims to develop an in vitro model to replace in vivo tumourigenicity studies for safety assessment of genome edited human haematopoietic stem cells (hHSCs).
Background
Diseases such as sickle cell anaemia, haemophilia, thalassemia and severe combined immunodeficiency result from modifications in a single gene occurring throughout the cells of the body. In the European and East Mediterranean area alone, at least 2,000 births a year are affected 1,2,3. The only curative treatment is an allogenic haematopoietic stem cell (HSC) transplant, which involves transferring HSCs from a healthy donor to the patient’s body after high-intensity chemotherapy or radiation. These treatments are expensive, (the cost of the transplant is ~ $275,000) 4, require patients to undergo lifelong pharmacological immunosuppression (costs ~ $12,000/year/patient 5, and appropriate donors need to be identified to prevent adverse immune responses.
A number of blood-related monogenic diseases have the potential to be cured with genome edited human HSCs (GE-hHSCs) that have been modified by inserting, deleting, modifying or replacing genetic material. This has created a significant amount of research and development to penetrate a market expected to reach $8.1 billion by 2025 6. Eight clinical trials based on the use of GE-hHSCs are ongoing (https://clinicaltrials.gov/), and the number is expected to increase over the next decade.
The ex vivo genome modification of hHSCs can be achieved using vectors to integrate genetic material into cells and the method is currently under development using designer nucleases 7. Designer nucleases, including but not limited to zinc finger nucleases, TALENs, and the CRISPR/Cas9 system, target one specific DNA sequence and create a double-stranded DNA break. Repair by non-homologous end joining can lead to mutations or, if a donor template is made available, gene replacement by homology directed repair. Designer nucleases offer a high level of precision but can cleave undesired genomic regions, causing off-target modifications, which could generate cells with functional impairment, altered fitness, or oncogenic potential 8. Recent studies have shown that human induced pluripotent stem cells (iPSCs) that survive CRISPR/Cas9-mediated editing present concomitant mutations in the P53 gene 9. Although the selection of p53 mutants does not appear to happen in hHSCs 10, other unexpected modifications could occur, and genome edited cells need to be broadly assessed for such safety liabilities prior to their use in the clinic.
Current assessment of potential off-target effects includes in silico prediction tools, extensive DNA analysis of in vitro edited cells/DNA (using GUIDE-seq, Digenome-seq, CIRCLE-seq, SITE-seq, BLESS/BLISS, HTGTS, IDLV capture and others) and in vivo carcinogenicity/tumourigenicity studies 8,11.
In silico analysis can predict, and DNA-based analysis can confirm, the presence of off-targets, but they cannot anticipate the functional consequences of those mutations. In vivo carcinogenicity studies, most often using NSG (NOD-SCID IL2Rγ−/−) mice, are lengthy, costly, require large numbers of animals and are poorly predictive of human safety, often due to poor engraftment of human cells in the animals 12,13, 14,15. Extending the study observation time has been shown to increase engraftment of cells, but extended studies present increased welfare burdens on the animals and costs 16. Engraftment rates have improved by generating NSG mice that express human cytokines 17,18,19,20 or by placing hHSCs within subcutaneously implanted humanised synthetic ossicles. While promising, these approaches still require animals and their use for tumourigenicity assessment of GE-hHSC has not been evaluated 21,22.
In vitro models to assess tumourigenicity are available but are currently not fit-for-purpose for the assessment of GE-hHSCs. They include:
- A soft agar colony forming assay has been shown to be more sensitive and predictive for assessing the tumourigenicity of iPSCs when compared to in vivo studies 23 (and unpublished data from HESI CT-TRACS). However, it is not appropriate for cells in suspension, such as hHSCs, that are intrinsically anchorage independent.
- A haematopoietic specific colony forming unit assay which is frequently used to evaluate toxic effects on HSCs is not optimised for tumourigenicity assessment.
- 3D co-culture systems that generate a humanised HSC niche exclusively in vitro, have been published 24,25,26,27. In these studies, only a single organ is present and the parameters to distinguish between normal and malignant hHSCs are not identified.
These emerging techniques provide a strong basis for the development of a more complex system suitable for animal free assessment of tumourigenic potential of hHSCs.
3Rs benefits
In vivo studies to assess the tumourigenicity of genome edited products are a regulatory requirement 28,29. These studies are lengthy, costly, require the use of large numbers of animals and are not always predictive of the risk to human safety 30.
Novartis currently runs at least one study per year on GE-hHSCs. A typical in vivo experiment requires a minimum of 200 to 300 immunocompromised mice and a follow up of at least 20 to 24 weeks. Mice are often irradiated, then dosed via an intravenous infusion, followed by repeated peripheral blood sampling to confirm engraftment which can cause stress and discomfort in the animals, and assessment for potential tumour formation. Engraftment is not always successful which means that many animals are dosed but do not provide useful data, potentially requiring repeat studies to be performed.
An in vitro assay developed through this Challenge could potenially replace these in vivo tumourigenicity studies. There is also significant potential for the assay to be used for the assessment of other gene therapy products such as viral-mediated gene transfer or antisense oligonucleotides, replacing the use of thousands of rodents in tumourigenicity assessment. The assay could also be used to identify appropriate dosing regimens and assess toxicity of new compounds impacting haematopoiesis.
Full Challenge information
Phase 2 winner
Phase 2 awarded to a team led by Dr Alessandro Rotilio, MOAB srla, a spin-off of Politecnico di Milano incubated by Hub-Initio, they have been awarded £1,000,000 to deliver Phase 2.
Read more about MOAB on their website.
Phase 1 winners
Phase 1 Project teams led by:
- Dr Uwe Marx, TissUse GmbH, Germany, £99,960.
- Professor Manuel Salmeron-Sanchez, University of Glasgow, £99,278.
- Professor Manuela Raimondi, Politecnico di Milano, Italy, £98,824.
Assessment information
Challenge Panel membership
| Name | Institution |
|---|---|
| Dr Martino Picardo (Chair) | Independent |
| Dr Silvana Libertini (Sponsor) | Novartis |
| Dr Marian Raschke (Sponsor) | Bayer |
| Dr Qin Wang (Sponsor) | Takeda |
| Professor Michael Capaldi | Newcastle University |
| Dr Neil Rodrigues | Cardiff University |
| Professor Amir Ghaemmaghami | University of Nottingham |
| Professor Dominique Bonnet | The Francis Crick Institute |
| Professor Dr Axel Schambach | Hannover Medical School |
| Professor Dr Peter Ertl | Technische Universität Wien (TUW) |
Through this Challenge, a multi-disciplinary team led by the Italian SME MOAB srl and supported by industry Sponsors Bayer AG, Novartis and Takeda, has developed a dual chamber bone marrow and lymph node bioreactor platform for assessing the tumorigenicity of gene-edited human haemopoietic stem cells (hHSCs). The MOAB is a milli fluidic bioreactor, composed of optically accessible chambers equipped with scaffolds to support 3D culture, which are connected and perfused.
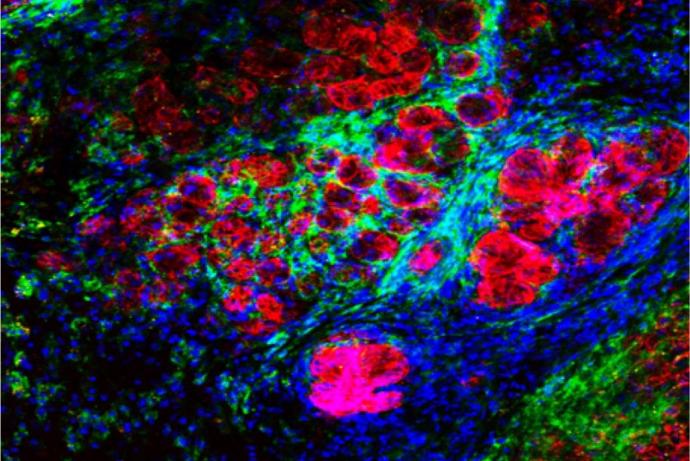
The team showed that hHSCs could be maintained in the bone marrow chamber for up to three months (Ritter P et al. 2024). Next, they tested the capability of the system to detect tumorigenic events. They first showed that by mixing hHSCs with engineered fluorescent tumour cells and following their growth that the platform could detect 500 tumorigenic cells in 100,000 hHSCs (limit of detection: 0.5%). Non-tumorigenic cells colonised the bone marrow model, whereas the tumour cells expanded and entered the perfusion. Lastly, hHSCs from multiple donors were edited using CRISR/Cas 9 technology to harbour a translocation known the drive leukemia development. Differences in the proliferation rates and differentiation between gene-edited and control cells were detected in the system. Gene-edited hHSCs that entered the lymph node developed a myeloid phenotype and showed an increase in proliferation compared to control cells.
The team has demonstrated the first proof-of-concept for assessing the tumorigenicity of gene-edited hHSCs in the MOAB bioreactor system. They are looking to test further gene-edited hHSCs using a range of gene-editing technologies to fully qualify the system.
For more information on the MOAB bioreactor, please contact Dr Manuela T. Raimondi (manuela.raimondi@moab-research.com).
Ritter, P. et al. (2024) ‘A millifluidic bioreactor allows the long term culture of primary lymphocytes or CD34+ hematopoietic cells while allowing the detection of tumorigenic expansion’, Frontiers in Bioengineering and Biotechnology, 12. doi:10.3389/fbioe.2024.1388312.