CRISPR/Cas9 technology
One or two, well-controlled and planned breeding steps can greatly enhance the reproducibility and robustness of phenotyping data produced from gene modifications using genome editing techniques.
The world of mammalian genetics has been set alight over the last 18 months with the discovery and introduction into many laboratories of RGEN (RNA-guided endonuclease) technologies, sometimes better known as CRISPR/Cas9 [1].
This technology comes with the promise that the genome of potentially any organism could be changed easily and efficiently. In rodents for example, this can be done by simply injecting one-cell fertilised embryos with a prokaryote-derived protein (e.g. Cas9) and one or more ‘guide’ RNAs which would direct the precise alteration of single or multiple genes [2].
In the cold light of day however, things aren’t that simple. Concerns regarding off-target effects where genomic alterations occur elsewhere in the genome (particularly in cell culture) and mosaicism of founder animals have been well discussed in many forums [4]. Modified proteins, altered RNA designs and different injection strategies are being investigated throughout the transgenic technology world to refine the accuracy and increase efficiency of these techniques. However, no refinements to date have managed to eliminate the creation of mosaics in the F0 generation.
This is a fast progressing field where many aspects will be discussed widely in meetings, on mailing lists and between collaborators. However, one discussion has reached a consensus amongst many transgenic scientists. That it is best practice NOT to use F0 animals for phenotyping and experimentation. The production of F0 mice homozygous for alterations in a single gene initially ignited the hope that little or no breeding would be required to produce a cohort of experimental animals [5]. This would both reduce animal numbers and costs. However, central to good scientific practice has to be the robustness and reproducibility of results. The major factor that makes the use of F0 mice for phenotyping inappropriate is the frequent occurrence of mosaicism in F0 animals.
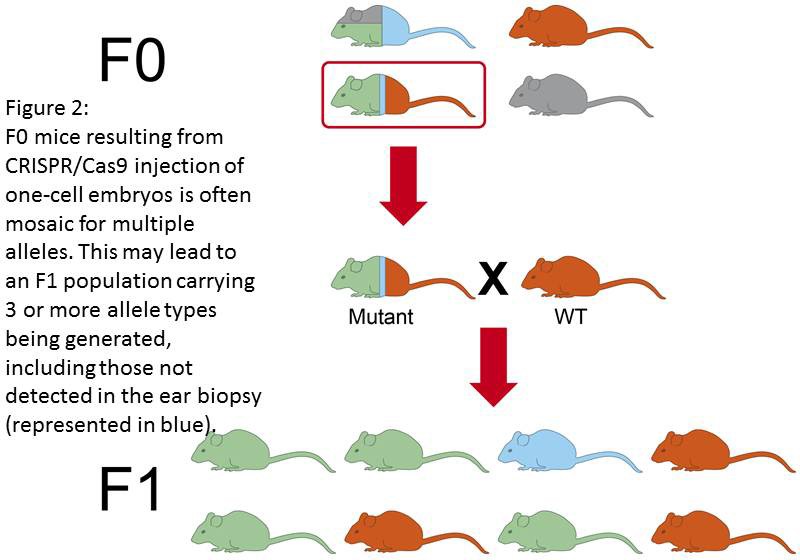
Genetic mosaicism occurs when action of the RGEN alterating the genome is delayed beyond the single-celled embryo. Nuclease-induced mutagenesis in embryos may occur slower than zygotic division. Therefore a situation may occur that gene modification is not achieved until the 2-, 4-, 8- or even later stage of embryo development (Figure 1). This may result in an animal being generated that contains a mixture of cells, some of which have the modification some of which don’t. Moreover, using CRISPR/Cas9 technologies can lead to DIFFERENT genetic alterations at the same gene loci, in different embryonic cells within the same animal. This may result in different cells of the mosaic animal carrying more than 2 different mutated allele types at the locus being targeted (Figure 2). Indeed it is possible for a single F0 animal to transmit multiple (5+) different alleles as its germ cells contain different populations of genetic alterations. Conversely, the desired genetic modification may not be transmitted to the F1 population.
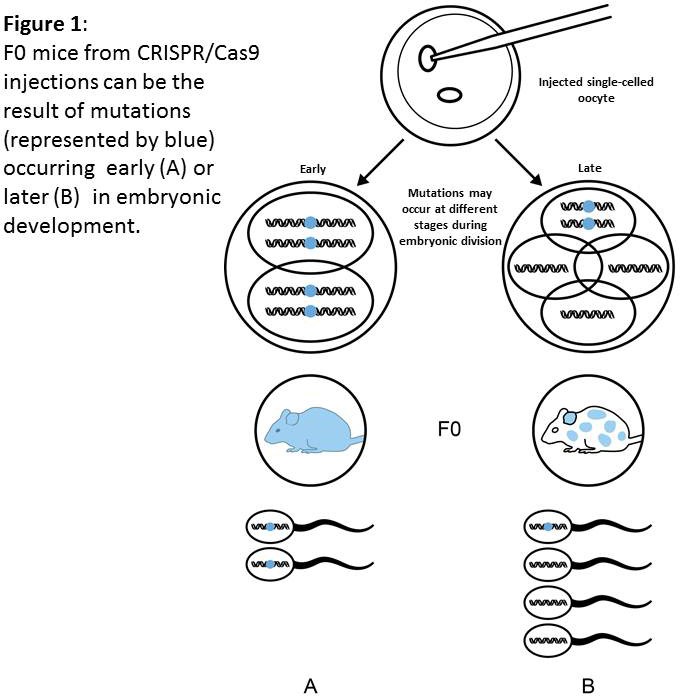
Clearly the data from phenotyping tests on such animals is neither reproducible (as the level of mosaicism is difficult to assess or control for) or, more importantly, appropriate for scientific interpretation.
There are also other factors that make the use of F0 animals for experimentation questionable, these include any maternal effects the strain used as foster mothers for F0’s may exert [3] and any effects of culture and microinjection. Both factors are difficult to control for and potentially add variability which will influence the integrity of scientific results.
References
- Hsu PD et al. (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157(6): 1262-78. doi: 10.1016/j.cell.2014.05.010
- Sander JD and Joung JK (2014) CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32(4): 347-55. doi: 10.1038/nbt.2842
- SenthamaraiKannan P et al. (2011) Identification of maternally regulated fetal gene networks in the placenta with a novel embryo transfer system in mice. Physiol Genomics 43(7): 317-24. doi: 10.1152/physiolgenomics.00078.2010
- Cho SW et al. (2014) Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 24(1): 132-141. doi: 10.1101/gr.162339.113
- Wang H et al. (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering Cell 153(4): 910-8. doi: 10.1101/gr.162339.113
Information on managing techniques for the generation and breeding of genetically altered mice, and strategies for improving welfare.

