Skin swabbing for DNA sampling of zebrafish
Learn more about skin swabbing as a welfare refinement, and how to establish a protocol for skin swabbing in your laboratory.
On this page
Introduction
Small bony fishes, such as zebrafish and stickleback, are commonly used as experimental models in the laboratory. DNA is routinely collected from these animals for genetic identification (genotyping). The current standard procedure to sample DNA is fin clipping, which involves placing the fish under nonterminal anaesthesia and removing a portion of the caudal fin with a scalpel. While fin clipping reliably generates good quality DNA samples for identifying animals by PCR, there is evidence that it affects fish health and welfare, leading to pain, stress and changes in behaviour [1-7, reviewed in 24]. This in turn can result in greater variation in physiological and behavioural data [8].
An alternative to fin clipping is skin swabbing, where a rayon-tipped swab is used to collect mucus from the flank of the fish, which can then be processed to extract DNA [9-11]. Behavioural and physiological evidence suggests that skin swabbing has a lesser impact on fish welfare [8,11], providing an opportunity to refine DNA sampling procedures for laboratory fishes. Read the FAQs for more on how the techniques compare from a fish welfare perspective. In terms of practicality, the swabbing method is comparable to fin clipping and has some potential advantages that are outlined in the table below.
Dr Will Norton and colleagues, at the University of Leicester and Aberystwyth University, have shown that skin swabbing can be used to successfully collect DNA from zebrafish and stickleback. Interest in skin swabbing as a refinement is growing, and researchers and technicians have an increasing number of questions about whether this technique is applicable to their work, and how to perform skin swabbing correctly. This resource contains detailed information to help research groups learn more about skin swabbing as a welfare refinement, and how to establish a protocol for skin swabbing in their laboratory.

Fin clipping and skin swabbing comparison table
| Traditional method: fin clipping | Refined method: skin swabbing | |
|---|---|---|
| Animal size | Larvae > 3.5 mm | Currently validated for juvenile fish ≥ 20 mm |
| Sample collection time | ~60 seconds to anaesthetise and fin clip (not including time for recovery) | ~30 seconds to restrain, swab and return to holding tank |
| Analgesia | Required before and after procedure [2] | Not required [8] |
| Anaesthesia | Surgical anaesthesia required, MS-222 (tricaine) commonly used | Not required |
| Extraction method | HotShot method, isopropanol extraction and commercial kits | HotShot method or isopropanol extraction; commercial kits are less effective |
| Quantity | Medium (ng/µl), suitable for PCR | Low (ng/µl), suitable for PCR |
| Cost | Cost per sample is comparable to skin swabbing (e.g. using HotShot method). Reusable scalpels or scissors may be used for multiple fish. | Cost per sample is comparable to fin clipping (e.g. using HotShot method). Consumable swabs are considerably cheaper than single-use scalpels; purchase of anaesthetic is not required. |
| Skill level | Requires training and individual authorisation; chemical restraint of the fish is necessary (anaesthesia) | Requires training; physical restraint of the fish is necessary
|
| Ethical approval | Licensed procedure requiring Home Office approval in the UK | Local ethical approval needed, e.g. from the AWERB, IACUC or equivalent |
Frequently asked questions
To answer your questions about zebrafish skin swabbing we spoke to experts in the swabbing technique, fish welfare and zebrafish genetics. Within the following FAQs the contributor is indicated by their initials (shown below).
- Dr Will Norton (WN), Associate Professor of Animal Biology at the University of Leicester
- Dr Gregory Paull (GP), Aquatic Facilities Manager and NACWO at the University of Exeter
- Karin Finger-Baier (KFB), Staff Scientist at Max Planck Institute for Biological Intelligence
- Professor Robert Kelsh (RK), Deputy Head of Life Sciences, University of Bath
WN: During recovery from fin clipping fish experience pain [2,4, reviewed in 24], which is not evident following the swabbing procedure [8,11]. During swabbing, fish will experience being netted and handled but the evidence suggests that this is less stressful than being anesthetised and recovering from the fin clipping procedure [8,11].
Part of the negative welfare effect of fin clipping comes from the necessary requirement to anaesthetise the animals. For example, MS222 (tricaine) treatment altered the opercular beat rate, a measure of ventilation in both zebrafish and sticklebacks [8] and commonly used anaesthetic agents have been shown to be aversive to zebrafish [17,18].
Swabbing is a less invasive procedure and does not require fish to be treated with analgesic or anaesthetic. By reducing the stress associated with the sampling procedure (i.e. avoiding anaesthesia and using a swab instead of a scalpel or scissors).
When fin clipping is used it is recommended that pain relief is provided to fish during recovery [2] and that the smallest amount of fin is removed as possible (ideally less than 10%) [24].
WN: Skin swabbing and fin clipping appear to be equally useful when amplifying genes to identify fish. DNA collection by skin swabbing tends to lead to a lower of concentration of DNA being extracted compared to fin clipping. However, our research has shown that the purity of the DNA is similar in both techniques [9].
We routinely use skin swabbing to identify zebrafish strains by PCR in our laboratory. We found that once we had familiarised ourselves with the skin swabbing sampling technique, we had very few samples that we could not amplify by PCR or sequence. We see a similar level of success when sequencing PCR reactions that used fin clip DNA as a template, but it may be that some practice is required to optimise the skin swabbing technique in your facility.
DNA from skin swabbing has been used to successfully identify fish by PCR in peer-reviewed literature [9-14]. The zebrafish strains included in these studies were AB wild type, casper, Tg(vmat2:GFP), Tg(hb9:GFP), Tg(glyt2:GFP) [9]; NHGRI-1, kca33Tg, and kca66Tg [10]. Sex markers have been also been used to differentiate between male and female stickleback [9], medaka [13] and wild roach [14] using DNA collected by skin swabbing.
We have not tried to use DNA extracted from skin swab samples for RAD-TAG sequencing or whole genome sequencing, so we cannot comment on the suitability of swabbing for these purposes at the present time. It is something that we would like to investigate in the future.
GP: You might find that swabbing is not suitable for all purposes, but if it works in 90% of cases that is already an improvement in terms of reducing the number of animals that go through this invasive procedure worldwide.
WN: Our research shows that the data collected from swabbed fish, which measured cortisol release, behavioural indicators of anxiety and gene expression, was less variable than in fin clipped fish. This creates an additional 3Rs benefit as fewer animals are required for sufficient statistical power. From a practical perspective, skin swabbing alone is not a licenced procedure (in the UK) and our laboratory personnel prefer using swabs to scalpels as there is less risk of harm to the user.
GP: There are also environmental benefits of not using anaesthesia; when anaesthetic agents are disposed of down the sink, they have the potential to affect ecological systems.
WN: Our full protocol is available to read online. You can get all the information you need on setting up your equipment and the practical steps of swabbing within this published methods article. To begin with, we recommend practicing restraining and releasing a fish with a hand net, as per the published protocol. It can be a little tricky to hold the fish the first time, but it gets easier the more you do it. As with all new techniques, it takes some practice to perfect your method, but this initial time investment is worth it to minimise the impact that DNA sampling has on fish health and welfare.
GP: I would recommend starting with large, short-finned adults as this will make handling the fish a little easier and will allow you to gain confidence in carrying out the DNA collection technique.
When it comes to carrying out the procedure, make sure you have your all your equipment ready and ensure that you only bring the number of animals to your procedure space that you can comfortably process without compromising their welfare. For example, fish should not be kept in off-system holding tanks long enough for there to be changes in water temperature.
KFB: My recommendation would be to start with lines with simple genetic makeups that produce strong bands when using DNA extracted from fin clips. Lines that show clear separation of fragments in gel electrophoresis, have a high yield of DNA product, do not require restriction digest to distinguish individuals by genotype and have no more than a single mutation or transgene will make a good starting point. When establishing skin swabbing, I would also recommend doing fin clips and swabs from the same line (not necessarily the same fish) in parallel and comparing the results (e.g. PCR, gel electrophoresis, sequencing etc.). Where these results indicate weak performance for a certain line (i.e. they generally give weak or barely visible bands during electrophoresis and/or have a low PCR yield) our solution has been to set up the PCR using twice the volume we usually use, and this has worked well.
KFB: Yes, some protocols might need to be adjusted slightly when switching to DNA obtained from skin swabs.
In our facility we had optimized our PCR settings (from fin clips) so that we could use a really small reaction volume and when we switched to skin swabbing we found that initially we did not get the desired results. To rectify this, we found that doubling the reaction volume was sometimes all that was needed.
Whenever we have complicated PCRs that have a high failure rate due to factors such as low gene expression, difficult DNA structure and formation of hairpins etc. we sometimes have to adjust the PCR parameters. In general, our PCR settings are chosen to give us the best possible result, starting with a line-specific DNA concentration. If starting with a lower concentration, the PCR settings might have to be optimized for good results again and just using the standard protocol might not be sufficient.
I would emphasize that one cannot assume that whatever the standard genotyping protocol for a given line is/was using fin clips, this can be copied one-to-one using skin swabbing. It might be necessary to make adjustments to your protocol when using DNA extracted from skin-swab samples, as we have done in our facility. However, in terms of refinement, this is absolutely worth the effort.
RK: Initially we struggled with high variability from swab samples but we have resolved this by making changes to how rapidly we prepare and process them. Since implementing this approach all the samples have given good DNA allowing for successful genotyping of all the genes tested so far.
Our approach is to take five swab samples, pause to begin the extraction process and then continue to collect another around of swabs. With two people we find that this method efficient, enabling seamless collection of samples.
WN: We restrain the fish using a combination of a hand net (e.g. a Marina Fine Soft Mesh Fish Net or similar) and wetted sponge with a groove cut into it (e.g. a Vitrex 10 2904 square sponge or similar). The thumb and forefinger can be used to hold the fish still, with the net covering the head and the tail of the animal (see the below image for an example of this). The fish’s body is placed into the groove on the sponge, which helps to keep it still. Some people prefer to just use a flat wetted sponge, with no groove [9], or single-use wet gauze rather than a sponge [10] – the technique that you find quickest and most practical will be down to your personal preference.
The aim is to take your sample and get the fish back into water as quickly as possible, so make sure that everything is ready to go before you net your fish out of the water. If you are not used to restraining small fish, we recommend practicing safe and efficient restraint before you have a go at swabbing. You’ll gain confidence quickly and will find what works for you.
GP: A common concern I hear is that handling smaller-bodied fish is challenging without sedation. If this is something you are struggling with initially, it may be advisable to start with slightly larger animals before progressing onto handling smaller fish once your confidence with the technique has grown.
To help the procedure run smoothly and to keep the fish calm, use a quiet space and avoid noisy rooms, vibrations and areas with high footfall. Fish can find exposure to bright lights stressful, which can influence how jumpy they are. Covering the holding container is often an effective way to reduce stress before you begin a procedure.
If you have practiced the technique and are taking reasonable steps to keep the fish calm handling unanaesthetised fish should become easier the more you do it. If you continue to struggle with manual restraint, for example, when handling a particularly hyperactive line, you may conclude that swabbing without sedation is not the best option for you. It is important to be open minded about what is possible with new techniques, but it is also essential to be pragmatic about what will work for your own situation. The onus is on us as researchers to refine our methods where possible; trialling skin swabbing will allow you to see whether it is a refinement appropriate for your individual circumstances.
RK: We simply use small nets and make sure the head is shaded to encourage calmness - I think covering eyes really makes a difference.
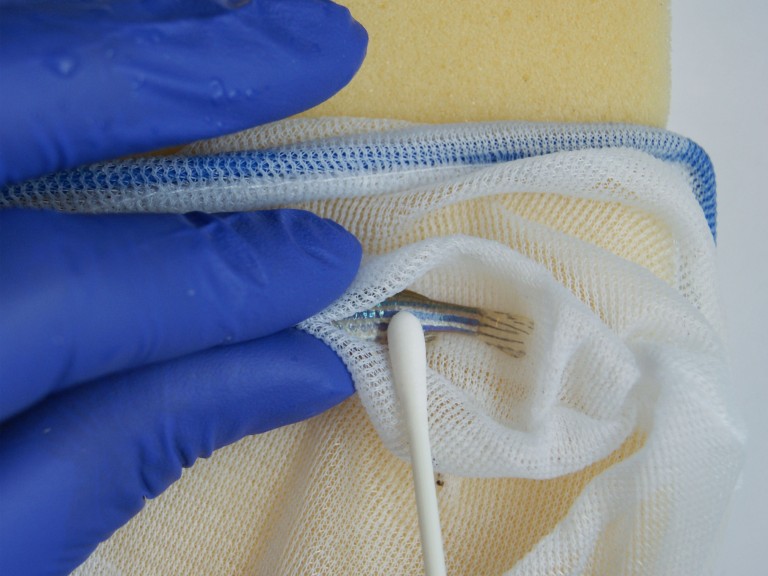
WN: Very little pressure is required when swabbing the fish, to prevent any possible discomfort or damage to the animal. The mucus can be collected readily by gently stroking the fish’s flank starting below the gills and moving towards the tail (this direction is important for fish with scales, such as zebrafish).
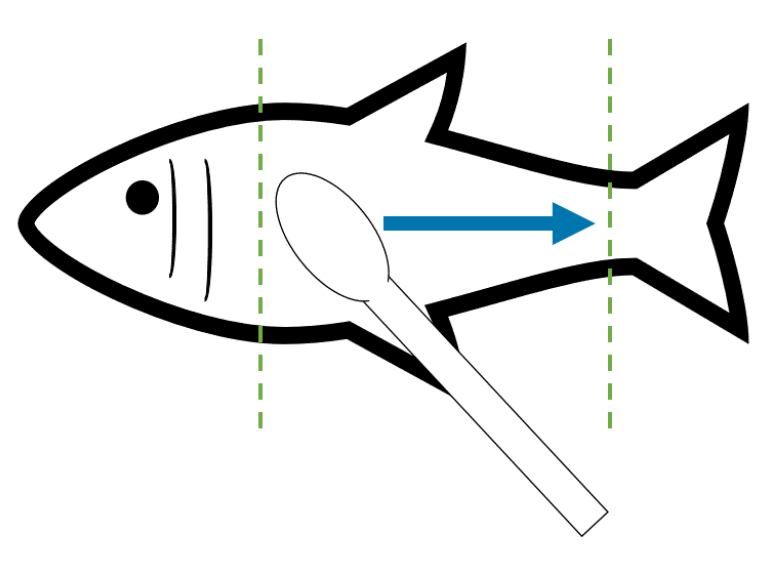
GP: Only light pressure is required, and the swab should be moved along the fish’s flank towards the tail in one clean movement. Avoid swabbing the operculum/gill area. I would suggest starting one swab width posterior of the gill plate, and even starting half way down the body will suffice if this means avoiding unnecessary stress by having to reposition the fish. On reaching the tail, lift the swab clear of the fish and repeat to retrieve enough material. For long-finned fish, you may need to gently clear the pectoral fin away from the body before collecting your DNA sample. This can be done by using the same swab.
WN: We have reliably collected DNA from fish ≥ 20 mm using the skin swabbing technique [9]. We are exploring whether swabbing zebrafish as small as ≥ 10 mm is viable, however this requires careful investigation and so we do not currently recommend the technique for fish smaller than 20 mm.
The Zebrafish Embryonic Genotyper (ZEG) is an option for genotyping zebrafish at the embryonic stage. Read about an aquarium technician's experience of using the ZEG in Tech3Rs (Issue 7, 3Rs Champions).
WN: We have successfully used commercially available rayon-tipped swabs. While these are consumable items they are easy to obtain for a low cost. We are hoping to trial the use of smaller-tipped swabs that can be used in 96-well plates.
WN: It does not matter if the swab gets a little wet when collecting mucus from the fish’s flank. This will not affect subsequent DNA extraction.
WN: Skin swabbing collects skin cells that have been shed by the fish and are trapped in the fish’s mucus layer.
WN: Studies show that cross contamination of this kind is not an issue [9,12]. We have demonstrated that cross contamination does not occur between zebrafish, even when the fish have been living at high densities and have been in direct contact with each other shortly before being swabbed [9].
WN: No, skin swabbing has been successfully used as less invasive method for DNA sampling of fishes with a body length ≥ 30 mm [12,13], amphibians [15] and mammals [16]. Our research has validated skin swabbing for DNA sampling in smaller-bodied (≥ 20 mm) laboratory fishes. Typically fin clipping is used as standard to obtain DNA samples from laboratory fishes and swabbing presents an opportunity for a refinement.
GP: My colleague at the University of Exeter, Dr Anke Lange, has successfully used skin swabbing for determining the genetic sex of roach (juvenile and adults) and stickleback. Within the same research group, they have also successfully used skin swabbing for metabarcoding the skin microbiome of tilapia [17].
WN: In our hands, we found that DNA extraction using standard molecular biology methods (phenol:chloroform extraction) worked better than commercial kits for skin swab mucus samples obtained from zebrafish and stickleback [9]. Other researchers have combined the skin swabbing protocol with a HotShot DNA extraction [10].
WN: We recognise that experienced personnel are able to carry out fin clipping efficiently and that facilities are committed to minimising harm to their animals, but we are only able to comment on what the data shows us. There is strong evidence that commonly used anaesthetics are aversive to fish [18-22] and that the fin clipping procedure itself causes stress and pain [1-7]. Skin swabbing is still a procedure that requires care; however, our data show a reduced impact on fish welfare [8] and we believe that this presents an opportunity to refine DNA collection in laboratory fishes.
Our end goal is improve the welfare of laboratory fishes and we invite others to share any data they have to highlight opportunities for refinement, either directly with myself (whjn1@le.ac.uk) or with the NC3Rs (tech3rs@nc3rs.org.uk).
WN: If you would like help with a training programme, we’d be happy to provide support (will.norton@le.ac.uk). In the meantime, use the most refined anaesthetic protocol in terms of safety and aversive properties [18-22], ensure that analgesia is used to manage pain before and after the procedure [2,20,21] and allow fish to recover in visual contact with other fishes [23] (e.g. by pushing tanks together so fish can see each other or by partitioning tanks so that fish are separated but have visual and olfactory contact).
GP: Also consider how much tissue needs to be removed from the fish for your purpose and do not take more of the fin than is needed. I have spoken to zebrafish users who have reviewed their fin clipping protocols and found that only a very tiny piece of tissue (e.g. 1-2mm2 equivalent to the tip of one tail lobe) was needed to complete >10 PCR runs. Fin clipping tail lobes back to the scissor of the tail was found to be completely unnecessary. The smaller the piece of tissue taken, the faster the recovery of the fish and sooner it can be returned to its home tank.
Protocol
See the Detailed skin swabbing protocol for information on how to carry out the swabbing technique to collect DNA from small-bodied fish species such as zebrafish. The protocol includes information on the equipment required and how to set it up, the preparation of reagents and DNA extraction.
You can also find useful tips for optimising swabbing within the FAQs on this page.
Video
Watch the below video to see how the swabbing technique is performed using zebrafish. The same method can be applied to collect DNA samples from other small-bodied laboratory fishes.
Research papers on skin swabbing of laboratory fishes
Tilley C et al. (2021). Skin swabbing protocol to collect DNA samples from small-bodied fish species [version 2; peer review: 2 approved]. F1000Research 10:1064 doi: 10.12688/f1000research.73115.2
Tilley CA et al. (2020). Skin swabbing is a refined technique to collect DNA from model fish species. Scientific Reports 10(1): 1-17. doi: 10.1038/s41598-020-75304-1
Venta PJ et al. (2020) A 13-plex of tetra-and penta-STRs to identify zebrafish. Scientific Reports 10(1): 1-7. doi: 10.1038/s41598-020-60842-5
Breacker C et al. (2017). A low-cost method of skin swabbing for the collection of DNA samples from small laboratory fish. Zebrafish 14(1): 35-41. doi: 10.1089/zeb.2016.1348
Le Vin AL et al. (2011). Validation of swabs as a non‐destructive and relatively non‐invasive DNA sampling method in fish. Molecular Ecology Resources 11(1): 107-109. doi: 10.1111/j.1755-0998.2010.02909.x
Acknowledgements
We gratefully acknowledge the contributions of Dr Will Norton (University of Leicester), Dr Ceinwen Tilley (University of Leicester), Dr Gregory Paull (University of Exeter), Karin Finger-Baier (Max Planck Institute for Biological Intelligence), Professor Robert Kelsh (University of Bath), Dr Chrissy Hammond (University of Bristol), Mollie Millington (The Francis Crick Institute), Dr Stewart Owen (AstraZeneca), and Dr Anke Lange (University of Exeter).
References
- De Lombaert et al. (2017). Behavioral characteristics of adult zebrafish (Danio rerio) after MS222 anesthesia for fin excision. Journal of the American Association for Laboratory Animal Science 56: 377–381. PMID: 28724486
- Schroeder PG and Sneddon LU (2017). Exploring the efficacy of immersion analgesics in zebrafish using an integrative approach. Applied Animal Behaviour 187: 93–102. doi: 10.1016/j.applanim.2016.12.003
- White LJ et al. (2017). The impact of social context on behaviour and the recovery from welfare challenges in zebrafish, Danio rerio. Animal Behaviour 132: 189–199. doi: 10.1016/j.anbehav.2017.08.017
- Deakin AG et al. (2019a). Automated monitoring of behaviour in zebrafish after invasive procedures. Scientific Reports 9(1):9042. doi: 10.1038/s41598-019-45464-w
- Deakin AG et al. (2019b). Welfare challenges influence the complexity of movement: fractal analysis of behaviour in zebrafish. Fishes 4(1): 8. doi: 10.3390/fishes4010008
- Thomson JS et al. (2019). Assessment of behaviour in groups of zebrafish (Danio rerio) using an intelligent software monitoring tool, the chromatic fish analyser. Journal of neuroscience methods 328: 108433. doi: 10.1016/j.jneumeth.2019.108433
- Thomson JS et al. (2020). Acute and chronic stress prevents responses to pain in zebrafish: evidence for stress-induced analgesia. Journal of Experimental Biology 223(14). doi: 10.1242/jeb.224527
- Tilley CA et al. (2020). Skin swabbing is a refined technique to collect DNA from model fish species. Scientific Reports 10(1): 1-17. doi: 10.1038/s41598-020-75304-1
- Breacker C et al. (2017). A low-cost method of skin swabbing for the collection of DNA samples from small laboratory fish. Zebrafish 14(1): 35-41. doi: 10.1089/zeb.2016.1348
- Venta PJ et al. (2020). A 13-plex of tetra-and penta-STRs to identify zebrafish. Scientific Reports 10(1): 1-7. doi: 10.1038/s41598-020-60842-5
- Tilley C et al. (2021). Skin swabbing protocol to collect DNA samples from small-bodied fish species [version 2; peer review: 2 approved]. F1000Research 10:1064 doi: 10.12688/f1000research.73115.2
- Le Vin AL et al. (2011). Validation of swabs as a non‐destructive and relatively non‐invasive DNA sampling method in fish. Molecular Ecology Resources 11(1): 107-109. doi: 10.1111/j.1755-0998.2010.02909.x
- Díaz C et al. (2019). Fast Multiplex real time PCR method for sex-identification of medaka (Oryzias latipes) by non-invasive sampling. MethodsX 6: 587-593. doi: 10.1016/j.mex.2019.03.011
- Lange A et al. (2020). A newly developed genetic sex marker and its application to understanding chemically induced feminisation in roach (Rutilus rutilus). Molecular ecology resources 20(4): 1007-1022. doi: 10.1111/1755-0998.13166
- Ward A et al. (2019). Skin swabs with FTA® cards as a dry storage source for amphibian DNA. Conservation Genet Resources 11: 309–311. doi: 10.1007/s12686-018-1018-z
- Okada M et al. (2017). An efficient, simple, and noninvasive procedure for genotyping aquatic and nonaquatic laboratory animals. Journal of the American Association for Laboratory Animal Science 56(5): 570-573. PMID: 28903830
- McMurtrie J et al. (2021). Relationships between pond water and tilapia skin microbiomes in aquaculture ponds in Malawi. bioRxiv. doi: 10.1101/2021.12.06.470702
- Readman GD et al. (2013). Do fish perceive anaesthetics as aversive? PLoS One. doi: 10.1371/journal.pone.0073773
- von Krogh K et al. (2021). Screening of anaesthetics in adult zebrafish (Danio rerio) for the induction of euthanasia by overdose. Biology 10(11): 1133. doi: 10.3390/biology10111133
- Collymore C (2020). Anesthesia, analgesia, and euthanasia of the laboratory zebrafish. In: The Zebrafish in Biomedical Research (Eds. Cartner S, Eisen J, Farmer S, Guillemin K, Kent M, Sanders G). 1st edition. Academic Press.
- Köhler A and Valentim AM (2022). Analgesia, anesthesia, and euthanasia in zebrafish. In: Laboratory Fish in Biomedical Research (Eds. D'Angelo L, de Girolamo P). 1st edition. Academic Press.
- Wong D et al. (2014). Conditioned place avoidance of zebrafish (Danio rerio) to three chemicals used for euthanasia and anaesthesia. PLoS One 9(2): e88030. doi: 10.1371/journal.pone.0088030
- White LJ et al. (2017). The impact of social context on behaviour and the recovery from welfare challenges in zebrafish, Danio rerio. Animal Behaviour 132: 189–199; doi.org/10.1016/j.anbehav.2017.08.017
- Sneddon LU et al. (2023). Pain management in zebrafish: Report from a FELASA Working Group. Laboratory Animals 58(3). 261-76. doi: 10.1177/00236772231198733

