3Rs Prize winners
Our annual 3Rs Prize competition recognises outstanding and original work in 3Rs research. Here we provide scientific summaries of the papers that have won the Prize since it started in 2005.
About the 3Rs Prize
Our annual international 3Rs Prize, sponsored by GSK*, is open to any researcher in academia or industry, in the UK of overseas. It recognises papers that describe outstanding and original work that has or could have major impacts on the 3Rs and can be awarded to the Principle Investigator, research lead or any other author.
Read more about the Prize: International 3Rs Prize.
*The award consists of a £28k Prize grant and a £2k personal award. A £20k contribution is provided by GSK with all remaining funds, including the personal award made by the NC3Rs.
3Rs Prize winners
- Validating non-animal derived antibodies
- Animal-free evaluation of hormone potency
- Organoid model of adaptive immunity
- Benchtop organ-on-a chip fabrication
- CNS barrier-forming cerebral organoids
- Poultry vaccine screening and 3D embryonic stem cell culture (joint winners)
- Stem cell model of motor neurone disease
- In silico cardiac drug safety trials
- Guidelines on laboratory rat housing
- Cerebral organoids and refined oral dosing for dogs (joint winners)
- Computer model of cardiac electrophysiology
- Establishing 3D liver organoids
- A breathing lung-on-a-chip
- Stem cell model of liver disorders
- Refined mouse handling techniques
- Animal-free medium for embryonic stem cells
- Retinal explant tissue culture system
- DNA fingerprinting for Schistosoma parasites
- Refining Leishmania infection in hamsters
- Natural routes of bacterial infection in mice
Validating non-animal derived antibodies

Prize winner: Dr Carl Laflamme, Dr Riham Ayoubi and Dr Peter McPherson, McGill University, Canada
An estimated $1BN in research funding is wasted each year in the US alone on ineffective commercial antibodies. Many fail to recognise the intended target protein in some or all laboratory applications or recognise additional targets and generate non-specific results. While it is difficult to estimate the total number of animals used in antibody production globally, one million animals per year are used for this purpose in the EU alone. To tackle the wastage of animals and resources in unreproducible research caused by poor antibody performance, the winning team of scientists created partnerships with academics, research funders and ten commercial antibody manufacturers, representing an estimated 27% of global antibody production. They agreed on optimised protocols to assess antibody specificity in the three most common uses of antibodies in biomedical research laboratories – Western blot, immunoprecipitation and immunofluorescence. Peter, Riham and Carl tested 614 commercially available antibodies provided by manufacturers to 65 target proteins involved in neurological diseases. The majority of target proteins were chosen by research funders and focused on neuroscience research, including those linked to Alzheimer's disease, Parkinson’s disease and amyotrophic lateral sclerosis.
Their scalable screening strategy involved a panel of cells expressing each target protein at sufficient levels to be detected by an antibody with typical binding affinity (1–50 nM). Multiple antibodies were tested against each target protein (an average of nine to ten per protein), including both animal-derived and non-animal derived reagents. The team could not identify a high-performance antibody for approximately a third of the target proteins, demonstrating the prevalence of poor binding affinity and target specificity among commercially available antibodies. For two thirds of the antibodies tested (409 out of 614), the team's characterisation data conflicted with the antibody supplier’s recommendations for use. As a result, the participating companies have withdrawn 73 poorly performing antibodies from the market and changed recommendations for 153 antibodies. Notably, recombinant non-animal derived antibodies performed better than traditional animal-derived reagents. Only one of the 65 proteins tested was not recognised by any of the non-animal derived antibodies tested, with the majority (49 proteins) identified by at least three different recombinant antibodies.
A lack of large-scale performance data comparing different antibody generation technologies has contributed to the slow uptake of replacement alternatives, despite the well-established reproducibility issues of traditional animal-derived antibodies. The authors characterisation dataset is available on ZENODO, a freely available data-sharing website operated by CERN. Based on their findings that recombinant antibodies performed better than monoclonal or polyclonal antibodies the authors encourage the increased generation of recombinant antibodies to replace the use of animal-derived antibodies.
To detect whether a primary antibody has bound to a target protein a secondary detection system is used. This secondary detection system (e.g. secondary antibody) is bound to a detectable molecule such as a fluorophore which allows for the quantification of the target protein. Currently standard secondary detection systems are not always compatible with the animal free protein binders used in recombinant antibodies. The team will use the £28K Prize grant to develop protocols for the optimisation of secondary detection systems that are compatible with recombinant antibodies. The team, who have been approached by manufacturers of animal-free protein binders, will conduct an independent comparison of their performance compared to animal-derived antibodies. The development of reliable secondary detection systems for recombinant antibodies aims to remove another barrier to their adoption to replace animal-derived reagents.
Ayoubi R et al. (2023). Scaling of an antibody validation procedure enables quantification of antibody performance in major research applications. eLife, 12. doi: 10.7554/eLife.91645.2
Highly commended: Dr Renata N.C. Nundes from the Oswaldo Cruz Foundation, Rio de Janeiro, Brazil
Currently, the only available treatment for snakebite envenomation is the administration of specific antivenom. Antivenoms are purified antibodies harvested from an envenomed animal – the potency varies between each batch and must be assessed as part of quality control testing. Potency determination relies on the murine lethality neutralisation assay as recommended by the WHO. In this test, antivenom therapies are mixed with lethal doses of snakebite venom and administered intravenously to mice. This can lead to severe suffering including paralysis, seizures, haemorrhage and death. Snakes of the Bothrops genus were responsible for 69% of snakebite events reported in Brazil in 2019 (almost 21,000 cases). Bothrops snakebites often cause permanent tissue damage, leading to limb amputation, disability and in some instances, life threatening complications such as tetanus and sepsis. In 2017, around 2,000 mice were used for Bothrops antivenom efficacy and batch release testing at the National Institute for Quality Control in Health in Brazil where this work was conducted.
Renata developed a cell-based cytotoxicity assay as an alternative to the murine lethality neutralisation assay specifically for Brazilian Bothrops antivenom potency testing. The in vitro method is based on the ability of Bothrops venom to induce cytotoxic effects in an immortalised cell line and assesses the capacity of the antivenom to inhibit this cytotoxicity. The in vitro assay provides a concentration-based measure of antivenom potency that allows direct comparison of their efficacy. This allows clinicians to make better informed treatment decisions, compared to the in vivo assay where the volume of antivenom that protects 50% of mice from death is measured and then used to calculate potency for clinical use.
The cytotoxicity-based in vitro method exhibits very good inter-assay and intra-assay reproducibility, as demonstrated through comparison with existing in vivo data in collaboration with three Brazilian antivenom manufacturers and the National Control Laboratory. Further work to define the regulatory acceptance criteria for these biological assays will allow for their wider uptake to replace the murine lethality neutralisation assay in antivenom production.
Nundes RNC et al. (2024). A Cytotoxicity Assay as an Alternative to the Murine Model for the Potency Testing of Bothrops jararaca Venom and Antivenom: An Intralaboratory Pre-validation Study. Altern Lab Anim 52(2):82-93. doi: 10.1177/02611929241237518
Animal-free evaluation of hormone potency

Prize winner: Dr Franesco Nevelli, Merck
Follicle stimulating hormone (FSH) is produced for fertility treatments, most predominantly for IVF to increase the number of eggs available to collect and fertilise. Over 70,000 IVF cycles were reported by the Human Fertilisation and Embryology Authority in the UK alone in 2021. FSH potency can be influenced by its purity and chemical and physical changes introduced during the manufacturing process and as a result each batch of FSH must be screened for its potency as part of regulatory requirements. Until recently this testing was conducted in the Steelman-Pohley bioassay, set out in the European and US Pharmacopoeias, (the collections of standards and quality control assays used by regulatory bodies). In the assay, female rats are injected daily with FSH and the ovaries are subsequently removed and weighed, with ovarian weight increase being used as a measure of potency.
The in vitro assay developed by Francesco and colleagues at Merck can replace the Steelman-Pohley bioassay and in 2022 was approved for use by the European Medicines Agency for two products used in fertility treatments. The assay consists of a human cell line, HEK-293, transfected to express the FSH receptor. Potency is assessed by measuring increased cAMP levels following FSH receptor activation. Comparative studies benchmarking the in vitro assay against the rat model demonstrated a very high level of concordance in a range of parameters essential to assessing potency, including distinguishing chemical and structural changes that affect FSH activity.
Regulatory bodies, including the European Medicines Agency, have approved the use of the in vitro assay in more than 70 countries. Francesco and colleagues are now working towards worldwide acceptance to enable Merck to fully replace all rats used for potency tests of Merck Healthcare marketed products, with estimated saving of nearly 40,000 animals annually, based on forecast animal-use in 2023. The prize grant will be used to fund educational programmes for physicians and patients in the USA to increase acceptance of in vitro quality control processes for biological products.
Nevelli F et al. (2023). Biological Assay to Determine Gonadotropin Potency: From In Vivo to In Vitro Sustainable Method. Int J Mol Sci. 24(9):8040. doi: 10.3390/ijms24098040
Highly commended: Dr Lorna Ewart, Emulate Inc
Drug-induced liver injury is a common reason why potential therapeutics fail in clinical trials despite preclinical screening in both in vitro and in vivo models. In this year’s highly commended paper, Lorna and colleagues describe a screening strategy with “Liver-Chips”, which helps to avoid the use of animals by identifying toxicities early in the drug development pipeline and therefore preventing compounds destined to fail in the clinic being tested in vivo. The Liver-Chips contain four human cell types with hepatocytes embedded in extracellular matrix in one channel and immune and vascular cells in a separate channel. Culturing the cells in the chips replicates key histological features of the liver, including cuboidal liver cell morphology and bile canaliculi-like structures, and stable expression of liver-specific genes.
To demonstrate the utility of the screening strategy, Lorna and colleagues analysed across a blinded set of 27 known hepatotoxic and non-toxic small molecule drugs the performance of the Liver-Chips, in the largest characterisation study of this kind to date. Liver-Chip screens had no false positives, and successfully identified over 80% of drugs known to cause drug-induced liver injury, outperforming the data obtained from animal studies. This study helps to build confidence in the utility of organ-on-chip technologies in the short-term to help minimise the use of animals in drug testing and in the longer term to replace in vivo testing with complex in vitro approaches.
Ewart L et al. (2022). Performance assessment and economic analysis of a human Liver-Chip for predictive toxicology. Commun Med. 2(154). doi: 10.1038/s43856-022-00209-1
Organoid model of adaptive immunity

Prize winner: Dr Lisa Wagar, California Irvine University
In Lisa’s winning paper, her entirely animal-free organoid model of the adaptive immune response provides new capabilities and insights to support the rapid development of broadly protective vaccines.
Research into adaptive immunity and vaccine responses still largely relies on animal models including mice, rabbits, hamsters, guinea pigs and pigs. Many vaccine candidates developed using animal models ultimately fail in human trials suggesting they fail to replicate the complex responses of the human immune system. Lisa’s tonsil organoids are an accessible alternative that replaces the use of animals and animal cells and reduces use of animal-derived products (such as cell culture media/sera or antibodies) with a model which is faster, cheaper and more human relevant.
As a lymphoid organ, tonsils are important for generating immune responses to infection or vaccination and serve as a reservoir of lymphocytes (B cells and T cells). Lisa collected tonsil tissue, discarded from tonsillectomy surgeries, as a readily available source of human lymphoid tissue to create immune organoids. Cells extracted from the tonsil tissue reaggregated into 3D immune organoids when grown in culture and formed structures that mimicked germinal centres with distinct B cell clusters. When exposed to immune stimulating agents, the organoids underwent an antigen-specific adaptive immune response which included somatic hypermutation, affinity maturation and B cell class switching. In addition to replicating these key aspects of the human immune microenvironment for the first time in vitro, Lisa’s model overcomes key barriers to existing replacement methods including the need for specialised equipment, challenging technical protocols and low throughput.
Lisa used her organoid system to investigate the immune response to vaccines for influenza, MMR, rabies and COVID-19, and how adjuvants and priming antigens can boost this response to increase vaccine efficacy. Following stimulation with live attenuated influenza vaccine, Lisa observed B cell maturation, antibody secretion and differentiation and activation of T cell populations. She was able to define the essential cellular components required to produce a productive flu vaccine response and capture individual donor variation to better understand how and why vaccine efficacy varies across the population. This means that the system can be used to assess immunogenicity and immune response variability before clinical studies to support the development of vaccines that confer protective immunity as broadly as possible. Her results highlighted important differences in murine and human immune responses and identified possible strategies for future vaccine design.
The prize grant will support early careers researchers in the Wagar lab to present their research at conferences, showcasing the utility of the model and driving its adoption alongside building the future skills base in non-animal organoid approaches. Future work will aim to capture additional features of the adaptive response including immune cell migration and further validate organoid cultures against human tissue responses in immunised patients. By further developing the technology to handle more samples in less time, Lisa aims to maximise the efficiency of respiratory disease research, streamline vaccine development and tackle global pathogens of concern, all whilst replacing the use of animals.
Wagar LE et al. (2021). Modeling human adaptive immune responses with tonsil organoids. Nat Med 21:125-135. doi:10.1038/s41591-020-01145-0
Highly commended: Professor Alex Frangi and Dr Ali Sarrami-Foroushani, University of Leeds
The highly commended paper reports the first in silico clinical trial for a medical device, exemplifying the potential of computer-based testing and trials to replace the use of animals and accurately predict the safety and efficacy of medical devices.
In silico trials can provide evidence to ensure only treatments with the highest likelihood of clinical benefit and lowest risk of complications progress to later-stage testing, avoiding the use of animals in preclinical safety and efficacy studies. In their paper, Alex and Ali presented the flow diverter (FD) performance assessment (FD-PASS) in silico trial, which reproduced and expanded on findings from clinical trials of endovascular devices to treat brain aneurysms.
Research into treating vascular diseases of the brain (e.g. aneurysm and stroke) typically uses large animals including pigs, dogs and rabbits. Between 2000 and 2015, more than 430 rabbits and 160 dogs were used in studies focusing on the flow-diverting stents that Alex and Ali modelled in silico. By modelling the treatment of intracranial aneurysms in virtual patients, they were able to replicate the findings from in vivo experiments that took years and over 600 animals to produce, in a matter of days.
In addition, Alex and Ali’s in silico model goes beyond what is possible in conventional animal and human trials. They were able to identify new risk factors for treatment failure and assess an individual patient’s risk of ischemic and haemorrhagic complications, supporting clinical decisions to be made on a case-by-case basis. The model’s virtual patient approach has broad potential applications across disciplines, for example in cardiovascular (heart valves and pacemakers) and musculoskeletal (hip and knee replacements) medicine. The paper adds to the body of reliable regulatory evidence that is critical to support the adoption of in silico testing and trials in the UK and internationally. It was a key driver in establishing the in silico medicine innovation network (InSilicoUK), a community of scientists, learned societies and SMEs brought together by Innovate UK to advance the use of in silico approaches which replace the use of animals and improve patient outcomes.
Sarrami-Foroushani A et al. (2021). In-silico trial of intracranial flow diverters replicates and expands insights from conventional clinical trials. Nat Commun 12:3861. doi:10.1038/s41467-021-23998-w
Benchtop organ-on-a chip fabrication

Prize winner: Dr Daniel Ferreira at the Instituto de Investigação e Inovação em Saúde, Portugal
Daniel and colleagues have adapted a method to enable organ-on-a-chip systems to be designed, developed and printed with commercially available equipment and materials.
Organ-on-a-chip systems are a promising tool for reducing the reliance on animal models in basic and applied laboratory research. The devices consist of cells and/or tissue grown in microfluidic chips, which are composed of a clear flexible polymer. The chips are designed to closely mimic human physiology of a specific organ and can be applied in many research fields including disease modelling, toxicology and safety studies, and personalised medicine. Multiple chips, replicating different organs, can also be connected for organ-organ communication studies, and researching the interface between specific cells and tissues.
While organ-on-a-chip systems hold a lot of promise, there have been barriers to uptake including the cost and time to manufacture the devices, which are often bespoke. In his publication, Daniel demonstrates how commercially available sheets of thin polymer can be used with a cutting plotter to construct each layer of a microfluidic chip. The chip can then be assembled on a glass microscopic slide.
To better replicate human physiology, organ-on-a-chip systems often integrate membranes, Daniel describes how these can also be incorporated using the method featured in his publication. For example, one method to enhance the functional response of cultured hepatocytes is to flow cell media continually over a PET membrane above the cells, which promotes undisturbed fluid flow. The fabrication method featured in Daniel’s publication allows a PET membrane to be added, using bench-top laboratory equipment in a number of hours. These chips are stable at physiological temperature and humidity enabling in vitro toxicity testing.
Daniel also describes how biomechanical cues can be included within his method for chip assembly. When a vacuum is applied to a microactuator chamber a flexible membrane in the chip stretches expanding the substrate the cells are cultured on with resulting surface expansion equivalent to that within the gut, lung and heart. Daniel validated using a chip with an integrated microactuator by culturing a human gastric epithelial cell line using a stretching pattern designed to mimic peristalsis. Cells grown in these conditions had an average epithelial height equivalent to that observed in normal stomach epithelium and expressed Mucin-1 in a similar pattern to normal gastric mucosa. These qualities surpassed those of cells grown in static conditions and those grown with flow alone.
The method described in the 3Rs Prize-winning publication enables organ-on-a-chip prototypes to be rapidly designed and assembled. The modularity of chip fabrication allows each aspect of the chip, such as robustness, cell culture maintenance and mechanical stretching, to be independently tested before new layers are added. Any alterations to the chip design can then be implemented and tested rapidly. Daniel will now use the prize grant to begin his independent academic career establishing a microfabrication lab. He will design an organ-on-a-chip platform as a proof-in-principle to study the molecular and cellular mechanisms that initiate hereditary diffuse gastric cancer using patient derived induced pluripotent stem cells.
Ferreira DA et al. (2021). Alternative to Soft Lithography for the Fabrication of Organ-on-a-Chip Elastomeric-Based Devices and Microactuators. Advanced Science 8:2003273. doi: 10.1002/advs.202003273
Highly commended: Dr Ben Newland, Cardiff University
The highly commended publication describes a method to induce focal lesions in tissue slices to create an ex vivo model of multiple sclerosis (MS). MS is modelled in rodents by immunising against myelin, inducing experimental autoimmune encephalomyelitis, which results in myelin damage similar to patients with MS. These studies are associated with a high level of suffering and the need for specialist husbandry and care.
Ex vivo slice culture methods have previously been developed but these induce demyelination across the full tissue, which is not representative of how MS develops in patients. In his publication, Ben describes a method using cryogels to apply a demyelinating agent to slices of mouse brain or spinal cord tissue. Similarly to global demyelination models, the focal application results in demyelination, microglial invasion and remyelination. However, this method results in focal lesions surrounded by healthy tissue, replicating how MS presents in patients. Both grey matter and white matter can be used in this model by using brain slices or spinal cord slices. Up to six brain slices and ten spinal cord slices can be isolated from each animal reducing the number of animals needed for studies of MS.
Eigel D et al. (2019). Cryogel scaffolds for regionally constrained delivery of lysophosphatidylcholine to central nervous system slice cultures: A model of focal demyelination for multiple sclerosis research. Acta Biomaterialia 97:216. doi: 10.1016/j.actbio.2019.08.030
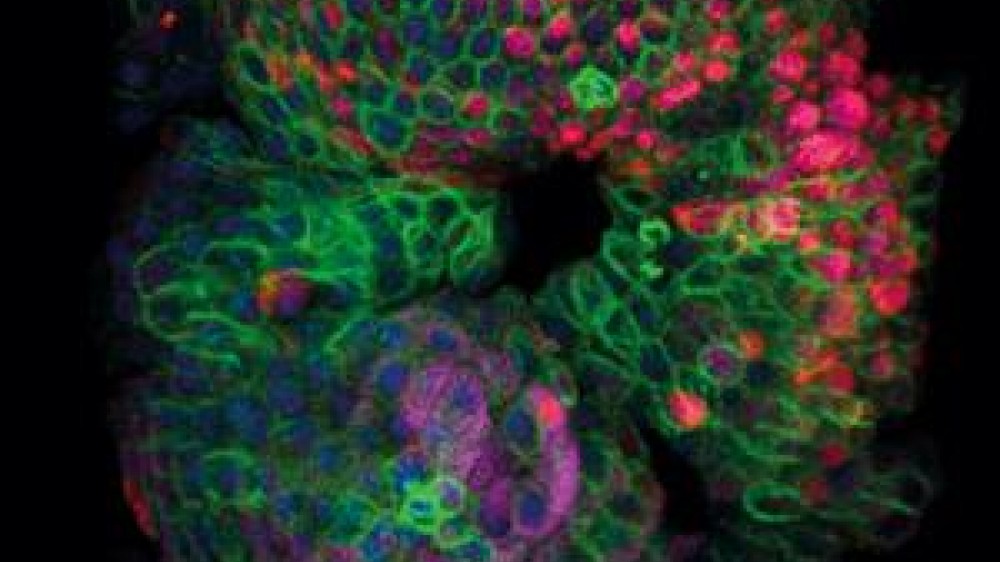
CNS barrier-forming cerebral organoids

Prize winner: Dr Laura Pellegrini, MRC Laboratory of Molecular Biology
The authors describe a method for developing cerebral organoids representative of the choroid plexus, the protective barrier between the blood and the cerebrospinal fluid (CSF), similar to the blood-brain barrier. The paper, published in Science in July 2020, describes a lot of firsts in in vitro neurological research, from accurately modelling the choroid plexus barrier function to their ability to produce and store cerebrospinal-like fluid.
Studying the choroid plexus is particularly challenging as the region lies deep in the brain, preventing patient biopsies from being used in research. Rodents and macaques are used to establish the ability of drugs to cross the choroid plexus and into the central nervous system, but many of these drugs fail in clinical trials. The choroid plexus has previously been modelled in vitro using organoid technology, but the lack of key features including the barrier function and cerebrospinal fluid production prevented their uptake to replace in vivo studies.
Introducing the human organoids described in the winning paper as an in vitro screening platform could help to prevent ineffective compounds progressing to in vivo studies. They can also be used to study the ability of pathogens to cross into the central nervous system, including the SARS-CoV-2 (COVID-19) virus. In addition, the cerebrospinal-like fluid produced by the choroid plexus organoids contains biomarkers and proteins similar to human cerebrospinal fluid, with each organoid yielding up to 20 times as much cerebrospinal-like fluid as a mouse embryo.
Highly commended: Dr Jennifer Ashworth, University of Nottingham
The highly commended paper by Dr Jennifer Ashworth and colleagues focuses on replacing Matrigel with non-animal-derived hydrogels. Matrigel is derived from mouse tumours and used in many in vitro studies to support cell growth. It is increasingly used in 3D cultures for lab-based models of disease, and approximately one hundred mice can be required to produce the amount of Matrigel needed by a single research institute every year. However, its chemical components are not well defined, affecting the reproducibility of studies
The hydrogels described in the highly commended paper, published in Matrix Biology, can be ‘tuned’ by adding proteins and sugars, altering their stiffness and composition to replicate different in vivo environments. The hydrogels have been used to support breast cancer cells, including cell lines and patient-derived samples, which would otherwise have needed to be sustained using large quantities of Matrigel, or by implanting into mice.
Poultry vaccine screening and 3D embryonic stem cell culture (joint winners)

Prize winner: Dr Francesca Nunn, Moredun Research Institute
Poultry red mites are a blood feeding ectoparasite. They are a global problem for the egg industry, affecting the welfare of laying hens through irritation and anaemia. Mites are controlled using chemicals, however, repeated use has led to resistance and recent research efforts have focused on developing vaccines and novel biopesticides. Assessment of vaccine control methods is initially done in vitro using blood assays before field trials are conducted, with 750 to 800 hens exposed to mites for each vaccine candidate, with an adjuvant control group. In vitro assays can be unreliable, for example, due to high levels of non-specific mite mortality. As a consequence, vaccine efficacy measured in vitro is not always translated into mite population reduction in field trials.
Francesca developed an “on-hen” mite feeding device that improves the screening of vaccine candidates to avoid unnecessary field trials. The device consists of a mesh pouch containing approximately 100 mites that have been starved for three weeks. The pouch is fitted to the thigh of the vaccinated hen – the mesh is large enough to allow the mites’ mouth parts to access the hen’s skin but small enough to contain the mites. Four hens are used per vaccine candidate and after three hours, the mesh is removed and the mites are recovered and maintained in 96-well plates for up to six days to assess mortality. The device has already been used to provide data that has prevented seven vaccines and vaccine delivery methods from going into field trials. The initial pre-screening using the on-hen device involved 56 hens in total, each exposed to 100 mites for three periods of three hours – the field studies would have used almost 5,500 birds exposed to 10,000 mites for 100 days. The on-hen device has been used by academic and commercial laboratories in the UK and internationally. By varying the size of the mesh, the device has the potential to be used in research on other parasites.
Prize winner: Dr Marta Shahbazi, MRC Laboratory of Molecular Biology
Implantation of an embryo into the uterus is a critical step with a high rate of pregnancies lost at this stage. Studying implantation and other early embryonic events is technically and ethically challenging. The majority of work is carried out in mice, typically genetically modified animals where associated surgery and breeding of large numbers of animals are required. Marta’s research has shown that it is possible to minimise this use with reproducible and novel 3D cultures of mouse embryonic stem cells that reliably mimic development at the time of, and beyond, implantation, avoiding the need for recipient mice for embryo transfer and the subsequent culling of animals to access early stage embryos.
Marta and colleagues have previously described an in vitro method to culture human embryos beyond the point of implantation, overcoming the technical challenges that have traditionally limited the use of human embryos in research. The winning paper builds on this by reporting comparative functional experiments using mouse and human embryonic stem cell 3D cultures that have identified key factors involved in the remodelling of the embryo at implantation to form the amniotic cavity. This has revealed a previously unknown link between cell potency and tissue shape, with a loss of stem cell “naïve pluripotency” (that is the ability to become any cell type in the organism) triggering the formation of the cavity and developmental progression of the embryo. The use of the 3D cultures for these studies replaced the use of 500 mice and importantly by demonstrating that they can be used to answer fundamental biological questions, the research has led to multiple groups worldwide adopting the cultures, further reducing the use of animals.
Stem cell model of motor neurone disease

Prize winner: Dr Rickie Patani, UCL Queen Square Institute of Neurology and the Francis Crick Institute
The authors used patient-derived stem cells to study motor neuron disease, also known as amyotrophic lateral sclerosis (ALS). They optimised a protocol for directed differentiation of induced pluripotent stem cells (iPSCs) into highly enriched (> 85%) functional populations of mature spinal cord motor neurons and astrocytes. Functional and transcriptomic analysis showed that the cells reflect key aspects of ALS pathology, including disrupted motor neuron synapse formation and mislocalisation of nuclear proteins leading to motor neuron death.
This model provides a tool to better understand the disease and to screen potential therapeutics, allowing some studies which are currently dependent on mouse models to be carried out in vitro. Dr Patani and colleagues have already transferred the robust iPSC differentiation protocols to collaborating academic centres, enabling three groups in the UK to shift some of their research from animal models. The cell model has a number of scientific benefits over the mouse models, which importantly include the ability to reflect the genetic heterogeneity of patients, investigate sporadic forms of the disease (90% of the cases), and allow early molecular events to be studied.
Hall CE et al. (2017). Progressive Motor Neuron Pathology and the Role of Astrocytes in a Human Stem Cell Model of VCP-Related ALS, Cell Reports 19(9): 1739-1749. doi: 10.1016/j.celrep.2017.05.024
Highly commended: Dr Diogo Mosqueira, University of Nottingham
This paper describes a human model of hypertrophic cardiomyopathy (HCM), generated by using CRISPR/Cas9 to introduce a disease-causing mutation in human pluripotent stem cells-cardiomyocytes (hPSC-CM). The authors produced 11 variants of mutation in a gene encoding the contractile protein beta myosin heavy chain, often affected in the disease. The cells were analysed as 2D monolayers and as 3D human engineered heart tissues, which partially recapitulate the architecture of cardiac tissue. Diseased cardiomyocytes showed hallmarks of HCM: hypertrophy and abnormalities in metabolism, contractility and calcium handling.
The cell lines are already being disseminated with three groups in Europe to further build confidence in the model and increase its uptake. In basic research and in early drug development, this cell-based method could be used to replace some animal models to examine the mechanisms of HCM and efficacy of candidate targets to treat HCM. In toxicology, the cells could be used to examine the potential for a compound to be harmful to people with HCM.
Mosqueira D et al. (2018). CRISPR/Cas9 editing in human pluripotent stem cell-cardiomyocytes highlights arrhythmias, hypocontractility, and energy depletion as potential therapeutic targets for hypertrophic cardiomyopathy. European Heart Journal 39(43), 3879-3892. doi: 10.1093/eurheartj/ehy249
Highly commended: Dr Bernhard Voelkl, University of Bern
The authors investigated how performing experiments in multiple laboratories affects the outcome of in vivo research, compared to experiments performed in single laboratories. They ran simulations based on 440 preclinical studies across 13 different interventions in animal models of stroke, myocardial infarction and breast cancer, comparing the accuracy of effect size estimates between single-laboratory and multi-laboratory study designs across a range of group sizes and experimental setups.
Single-laboratory studies generally failed to predict treatment effect accurately, resulting in an under- or overestimation of animal group sizes. Using more animals in single-laboratory studies often resulted in even less valid estimates of effect size, as this reduced variability between measurements but did not improve the accuracy of the measurements. By contrast, multi-laboratory designs increased accuracy by up to 42% without the need to use more animals. These findings demonstrate that, despite common assumption, within-study standardisation is a major cause of poor reproducibility.
In silico cardiac drug safety trials

Prize winner: Dr Elisa Passini, University of Oxford
The authors developed an in silico model that predicts the risk of drug-induced heart arrhythmias more accurately than animal studies.
They performed an ‘in silico drug trial’, testing 62 drugs and reference compounds at different concentrations on a control population of 1,213 simulated human ventricular cells.
Drug-induced changes in heart electrophysiology were measured in a user-friendly software, ‘Virtual Assay’, developed for this purpose and already used by four pharmaceutical partners. The computer model showed 89% accuracy in predicting the risk of drug-induced heart arrhythmias in humans, in comparison to up to 75% accuracy showed by data obtained from previously conducted animal studies.
Importantly the model also has the advantage that it represents the electrophysiological variability seen among patients. It outperforms other in silico approaches that average experimental data with a one-model-fits-all approach, and do not take inter-subject variability into account. This consideration of variability in simulations is crucial to capture differences in drug responses on a human population level and holds potential for identifying sub-populations at higher risk.
Highly commended: Dr Christian Tiede, University of Leeds
The paper shows that it is possible to generate Affimers, ‘alternative binding proteins’ to numerous targets, validating the use of antibody alternative binding reagents in molecular and cellular research studies. They isolated Affimers against 12 different targets and compared them to equivalent antibodies across seven different case studies. The researchers showed that Affimers, because of their specificity and other factors, can be equal to, or better than, antibodies for various molecular and cell biology applications.
The ability to create Affimers in the laboratory without the use of animals means they could potentially replace (animal-made) antibodies in a number of common molecular and cellular applications.
Highly commended: Dr Michael Walker, University of Guelph
The authors describe an alternative experimental design and analysis method – termed a ‘split-plot’ design – where animals are housed in mixed-strain groups. Female C57BL/6 (black), DBA/2 (brown) and BALB/c (white) mice were allocated to either enriched or standard treatments and screened for commonly measured and/ or welfare-relevant behavioural, physiological and haematological variables including corticosterone metabolite output and stereotypic behaviours. Results showed that living in mixed-strain trios did not reduce mouse welfare, or cause any change to strain-dependent or enrichment typical differences, validating it as an alternative study design.
The application of a ‘split-plot’ design meant fewer animals were required in each treatment group. The authors estimate, compared to a traditional design, this design can cut animal numbers required to achieve 80% power by more than half. Testing multiple strains increases the external validity of the experiment, thus increasing the generalisability of the results to other environmental contexts or populations.
Walker M et al. (2016). Mixed-strain housing for female C57BL/6, DBA/2, and BALB/c mice: validating a split-plot design that promotes refinement and reduction. BMC Medical Research Methodology 16:11. doi: 10.1186/s12874-016-0113-7
Guidelines on laboratory rat housing

Prize winner: Dr Joanna Makowska, University of British Columbia
The authors provided evidence that natural behaviours such as burrowing and standing upright are important for the welfare of laboratory rats. They observed the behaviours of rats in two different cage types: standard laboratory caging and large, semi-naturalistic cages with a dedicated area for burrowing.
They found that throughout their lives rats burrow readily, even when tunnels are already present, re-organising the tunnels on an almost daily basis. When allowed the space in large cages, rats stand upright multiple times per day, even until relatively old, and climb often when young. In comparison, rats housed in standard laboratory caging are unable to carry out these behaviours, instead stretching laterally, suggesting that they are compensating for their inability to stretch in the natural upright position.
This work provides a scientific basis for a change in guidelines on laboratory rat housing, including increasing cage height and providing burrowing materials. A number of laboratories have already modified their rat housing to accommodate some of the features described in the paper.
Makowska IJ, Weary DM (2016). The importance of burrowing, climbing and standing upright for laboratory rats. Royal Society Open Science 3: 160136 doi: 10.1098/rsos.160136
Cerebral organoids and refined oral dosing for dogs (joint winners)

Prize winner: Dr Madeline Lancaster, MRC Laboratory of Molecular Biology
The authors have developed the first 3D model of the human embryonic brain, using human induced pluripotent stem cells which were able to spontaneously self-organise into a structure that resembles the human brain with discrete, interdependent regions. This was achieved using adapted growth conditions, with specialised matrix support and improved access to nutrients through spinning. The paper also describes using skin cells from a patient with microcephaly to create organoids modelling the disease.
Suitability of animal models in studying neural development is limited, as they do not recapitulate the anatomical and functional complexity required to study human brain biology and disease. Developing brain organoids from human tissue is a revolutionary step towards reducing reliance on animals in studying neurological diseases and potentially in the development of new treatments.
Lancaster M et al. (2013). Cerebral organoids model human brain development and microcephaly. Nature 501(7497): 373-9 doi: 10.1038/nature12517

Prize winner: Dr Laura Hall, University of Stirling
The study, in collaboration with AstraZeneca, improves the technique of oral dosing in dogs. Following a framework of objective welfare assessments developed by Dr Hall, the authors demonstrate that a modified, refined protocol can minimise stress in dogs compared to the standard approach.
Most laboratory dogs in the UK are used for safety testing, and oral dosing is one of the most common procedures used during these tests. This paper shows that using seemingly small refinements (positive reinforcement training with food rewards, a signal for dosing, and covering the dosing tube in palatable paste during training) significantly reduces the negative welfare impact of the oral dosing on the dogs. The improved protocol allows researchers to dose dogs more quickly and efficiently, at no further cost. Dr Hall has been collaborating with dog facilities across the UK to maximise the impact of her work and share best practice.
- NC3Rs blog: Improving the welfare of laboratory dogs.
Hall LE et al (2015). Refining dosing by oral gavage in the dog: A protocol to harmonise welfare. Journal of Pharmacological and Toxicological Methods 72: 35-46 doi: 10.1016/j.vascn.2014.12.007
Highly commended: Dr Hayley Francies, Wellcome Trust Sanger Institute
The paper describes work on developing and characterising a biobank of colorectal cancer organoids obtained from patients’ biopsy tissue. The organoids were shown to accurately reflect the molecular features and diversity seen in the original tumours. More than 80 commonly used or experimental drugs for cancer were tested in the biobank.
Tumour-derived organoids have the potential to replace many studies involving patient-derived xenograft (PDX) mice, including for personalised medicine and drug development. Organoid production is cheaper and has a higher success rate than patient tumour sample engraftment rates in mice. This technology may fill the gap between cancer genetics and patient trials, complementing and in the long run replacing xenograft based drug studies, and allowing personalised therapy design.
Computer model of cardiac electrophysiology

Prize winner: Mr Oliver Britton, University of Oxford
The authors have built a computer model of cardiac electrophysiology that incorporates natural variability. Normally when using a computer model to test how a drug might affect the heart, the effect of the drug on the heart is compared to an average profile of electrophysiology. But this average profile is not really representative of the whole population, where natural variations in heart properties occur from person to person. This new approach has the potential to make computer models that are far more powerful and more predictive of human response, and therefore a more viable alternative to using animals in research. This is the first time that natural variability has successfully been considered in such a model, and the methodology could be applied to other diseases. The methodology has also been developed into a user-friendly software package called Virtual Assay, which is a major facilitator for industry uptake, without the need for specialist programming and modelling experience. The authors are already planning to use the same methodology to build computer models for understanding pain and diabetes.
Highly commended: Dr Olivier Frey, ETH Zurich
Frey’s paper reports on a novel approach to culturing multi-cellular spheroids in vitro. The work is a significant advance in engineering, which brings together the hanging drop method and microfluidics to substantially expand the experimental options for culturing spheroids. Growing cells using a hanging drop approach, means that 3D cell spheroids can be grown without the restriction that may be imposed by a scaffold or a dish. Microfluidic systems allow precise liquid handling including continuous medium and waste exchange, and also allow test substances, such as candidate drugs, to be run through the culture. This is the first time these two approaches have been combined, and the result holds real promise to bolster the predictivity of in vitro research. The microfluidic hanging drop network has already shown that spheroids of cells representing different organs can be inter-connected in physiological order and are able to communicate with each other via metabolite transfer. This is exciting as this capability is one of the first steps towards creating a multi-organ body-on-a-chip model.
Highly commended: Dr Nicola Powles-Glover, AstraZeneca UK
It is a requirement in animal safety testing of new medicines that the blood concentration of the medicine is measured. Historically, large volumes of blood were required to detect the concentration of the medicine. This meant that for rat studies, separate groups of rats were used solely for measuring the concentration while other groups of rats were used to assess the effects of the medicine on the animal. Advances in the way that blood is analysed mean that, with the right analysis equipment, very small “microsamples” of blood are now sufficient. Taking a microsample of blood from a rat is a quicker, less stressful procedure than taking a larger volume. This paper provides the evidence that taking repeat microsamples does not adversely affect adult rats and therefore does not interfere with the ability to interpret these safety studies. This means that information about the drug concentration and its affects can be obtained from the same animal, allowing a direct link between drug concentration and effect. This is a major scientific improvement. It also means that far fewer rats are required on these safety studies. As a consequence of this work, whenever the sensitive analytical method is available, AstraZeneca now use microsampling routinely in all rat safety tests.
Highly commended: Drs Brianna Gaskill and Joseph Garner, Purdue University and Stanford University
Mice are commonly housed at temperatures (20–24°C) which humans find comfortable for working in the laboratory. Mice become cold stressed below 30˚C, which can compromise many aspects of physiology and welfare. However, the amount of nesting material required to meet a mouse’s thermal needs has until now been unknown. In this study the authors found out where mice wanted to spend their time, based on combinations of temperature and nesting material. They found that mice prefer temperatures between 26–29°C, but shift from preferring a warmer temperature to a nest when provided 6-10g of nesting material. These results suggest that laboratory mice should be provided with no less than 6g of nesting material in order to build fully formed nests, but 10g or more may be needed to eliminate thermal stress. These results have the potential to positively impact the welfare of millions of laboratory mice all over the world.
Establishing 3D liver organoids

Prize winner: Dr Meritxell Huch, Gurdon Institute, University of Cambridge
Dr Meritxell Huch from Cambridge University's Gurdon Institute wins the 2013 3Rs Prize for a Nature paper detailing work carried out at the Hubrecht Institute, The Netherlands, to develop a culture system that enables adult mouse stem cells to grow and expand into fully functioning three-dimensional liver tissue.
Growing hepatocytes (liver cells) in the laboratory has been attempted by liver biologists for many years, since it would reduce their reliance on using mice to study liver disease and would open up new opportunities in medical research and drug safety testing. Until now no laboratory has been successful in deciphering how to isolate and grow these cells.
Liver stem cells are typically found in a dormant state in the liver, only becoming active following injury to produce new liver cells and bile ducts. Dr Huch and colleagues located the specific type of stem cells responsible for this regeneration, which are recognised by a key surface protein (Lgr5+) that they share with similar stem cells in the intestine, stomach and hair follicles.
By isolating these cells and placing them in a culture medium with the right conditions, the researchers were able to grow small liver organoids, which survive and expand for over a year in a laboratory environment. When implanted back into mice with liver disease they continued to grow, ameliorating the disease and extending the survival of the mice.
Having further refined the process using cells from rats and dogs, Dr Huch is now moving onto testing it with human cells, which would not only be more relevant to research into human disease, but also translate to the development of a patient's own liver tissue for transplantation.
Commenting on the new method's potential to reduce animal use in liver research, Dr Huch said:
"Typically a study to investigate one potential drug compound to treat one form of liver disease would require up to 50 live animals per experiment, so testing 1000 compounds would need 50,000 mice. By using the liver culture system I developed, we can test 1000 compounds using cells that come from only one mouse, resulting in a significant reduction in animal use.
Growing functioning liver cells in culture has been the Holy Grail for liver biologists for many years, so a limitless supply of hepatocytes could have a huge 3Rs impact both on basic research to understand liver disease and for the screening and safety testing of pharmaceuticals.
Highly commended: Dr Gyorgy Fejer, Plymouth University
Recognised for the development of a new method to grow macrophage cells for use in infectious disease research, which would reduce the use of mice by many thousands.
Highly commended: Dr Daniel Adams, University of California San Francisco
Dr Adams has taken inspiration from human orthopaedics to develop a biocompatible, titanium skull implant to reduce infection risk and improve welfare in monkeys undergoing cognition studies where brain activity is monitored directly.
Adams DL et al. (2011). A watertight acrylic-free titanium recording chamber for electrophysiology in behaving monkeys. J. Neurophysiol 106:15811590. doi: 10.1152/jn.00405.2011
A breathing lung-on-a-chip
Prize winner: Professor Don Ingber, Wyss Institute, Harvard University
Professor Ingber's research, published in Science Translational Medicine, describes an innovative 'lung-on-a-chip' microdevice that can accurately replicate conditions in a diseased human lung, offering a viable alternative to using animals in preclinical drug testing.
The microdevice contains hollow channels lined with living human cells, mimicking both the interface between tissues and the unique physical environment seen in whole living organs. Crystal clear and flexible, it is approximately the size of a USB memory stick.
Applying a vacuum to part of the microdevice allows it to 'breathe', recreating the way in which our tissues physically expand and contract during respiration. In testing it was able to successfully replicate the conditions seen in pulmonary oedema (fluid accumulation in the lungs), and predict results of a new drug for this life-threatening condition, which showed benefit in animal studies.
In addition, the microdevice has allowed the researchers to carry out real-time high resolution imaging on the cells and make accurate measurements of fluid flow and blood clot formation, which are not easily available in an animal model.
Professor Ingber said: "This is precisely the kind of progress that regulatory government agencies, such as the Food and Drug Administration (FDA) in the US, and pharmaceutical companies need to see in order to seriously consider an alternative approach to animal models."
In their paper the researchers describe how the next step is to apply the technology to other human organs with the goal of one day being able to use it as part of an automated system to test many drugs. While it is not expected to offer an immediate replacement for animal studies, further development and applications of the technology could allow for a more gradual replacement of animal models of human disease.
How the lung-on-a-chip works:
- Inside the microdevice are two parallel, sub-millimeter sized, hollow channels which are separated by a thin, flexible, porous membrane. This membrane is coated with matrix proteins that normally hold cells together in human tissues.
- One side of this membrane is lined with living human cells isolated from the air sac of a lung, and air is allowed to permeate into the channel to recreate the environment seen in a lung. The other side contains human lung capillary blood cells with a blood-like solution flowing over their surfaces.
- A vacuum applied to side chambers alongside the channels recreates the way our tissues physically expand and retract when we breathe.
- Recreating these conditions has been an important step to develop new insights into human lung disease that are difficult to achieve in with animal studies, such as the ability to carry out high-resolution imaging on the cells themselves, observing blood clot formation and fluid flow.
Highly commended: Professor Susan Barnett, University of Glasgow
Scottish-based researcher Professor Susan Barnett was commended for research developing an in vitro model of spinal cord injury using rat embryonic spinal cord cells. This has enabled the laboratory to test the combination of drugs being studied using cells from one animal only, representing a 97% reduction had an established methodology been used. This method is being further developed for testing therapeutics more widely.
Highly commended: Professor Gareth Sanger, Queen Mary, University of London
London-based Professor Gareth Sanger was commended for research demonstrating the benefit of using human - rather than animal - gastrointestinal tissues for drug testing, which are obtained as part of normal surgical procedures.
Highly commended: Professor Shuichi Takayama, University of Michigan (USA)
US researcher Professor Shuichi Takayama was commended for developing a 3D cell culture to test anti-cancer drugs, which proved to be more representative of clinical responses than standard 2D 'flat' cell cultures, demonstrating the potential for this method to replace and reduce the use of animals in pharmaceutical testing.
- NC3Rs blog: From 2D to 3D, not just a revolution on our TV
Stem cell model of liver disorders
Prize winner: Dr Ludovic Vallier, University of Cambridge
Dr Vallier's paper looks at the use of artificial liver cells to model inherited metabolic disorders of the liver has the potential to reduce the number of animals used in this type of research.
The artificial cells, known as human induced pluripotent stem cells (hIPSCs), offer possibilities to regenerate damaged tissues and organs, and it is their potential to reduce the number of animals used for screening potential drug treatments that led to Dr Vallier receiving the 3Rs Prize in 2011.
Human liver cells (hepatocytes) cannot be grown in the laboratory and differences between rodents and humans mean that it is rarely possible to recreate the human disease completely in mice or rats or to use cultures of rat or mouse liver cells. Dr Vallier's team took skin cells (dermal fibroblasts) from seven patients with a variety of inherited liver diseases and three healthy individuals (the controls). They then reprogrammed cells from the skin samples back into stem cells. These stem cells were then used to generate liver cells which mimicked a broad range of liver diseases and to create 'healthy' liver cells from the control group.
Ludovic Vallier's innovative study describes the development and validation of a method to produce cells similar to those in a human liver. Such cells could replace animals for some types of early drug testing and could also help us to predict adverse clinical reactions. Using these cells for drug testing could be transformative. Ludovic and his colleagues have well illustrated how addressing the 3Rs converges with improving the quality of science.
Highly commended: Dr Anna Williams, MRC Centre for Regenerative Medicine at the University of Edinburgh
Dr Williams' paper describes a new cell culture method that dramatically reduces the numbers of mice needed to test potential treatments for nerve cells damanged by multiple scleosis.
In multiple sclerosis (MS), immune cells enter the brain and cause inflammation and demyelination, where the protective covering of myelin around nerves is damaged. The brain can repair this damage by a process called remyelination, but this is not very efficient and frequently fails. Researchers have used rats and mice to try reproduce demyelination and to test for possible medicines to help promote remyelination. These experiments use large numbers of animals.
Dr Williams discovered that slices of brain taken from very young mice and grown in a dish, retain the three-dimensional structure and normal cells of the brain and can be used to test the effectiveness of medicines that might improve remyelination. The researchers also automated the ways in which they measured the remyelination, such that analysis now takes seconds rather than hours.
Highly commended: Dr Stephen Pettitt, Wellcome Trust Sanger Institute
Research conducted by Dr Pettitt and colleagues at the Wellcome Trust Sanger Institute has provided a new way of generating GM mice which avoids using two different strains and therefore reduces the number of animals used.
Scientists use genetically modified (GM) mice to study a range of diseases. But producing GM mice can be a lengthy process involving large numbers of animals. For technical reasons scientists often use a particular strain of mice for the early parts of the process. However, they then need to breed the mice with another strain to give animals with the desired genetic background for their experiments.
The 'Black 6' strain of mouse (so-called because of its coat colour) is preferred for many experiments. However, for years GM mice could only be made efficiently in a different strain, '129', from which it was easy to derive embryonic stem cells. This meant the GM 129 mice had to be bred with Black 6 mice for several generations (backcrossing) before the mutation could be analysed on a Black 6 genetic background.
This technique has the potential to reduce the numbers of mice used by hundreds per project. The cells are already in use in an international project to investigate all 20,000 mouse genes, and this can now be done with increased efficiency and fewer animals.
Dr Pettitt's work consisted of deriving embryonic stem cells the starting point for making a GM mouse directly from Black 6 mice, thus removing the need for backcrossing. Importantly, the cells were modified so that they could be easily seen by a difference in coat colour in the mice. This means that mice with the highest proportion of GM cells can be selected for breeding ensuring that only those animals likely to transmit the genetic modification to their offspring are used.
Refined mouse handling techniques
Prize winner: Professor Jane Hurst
Professor Jane Hurst's research has shown that a new way of handling laboratory mice can improve their welfare and the quality of the science they are used for.
Laboratory mice are usually picked up by their tails. Professor Hurst's study proves this method of handling causes high levels of anxiety and stress which can influence the outcome of experiments. By simply catching the mice using a plastic tunnel or cupped hands anxiety can be greatly reduced. This small change can be easily applied and has the potential to make a big difference to the welfare of every mouse used for research.
Animal-free medium for embryonic stem cells
Prize winner: Dr Jenny Nichols
Dr Jenny Nichols developed an optimised culture medium for growing mouse embryonic stem (ES) cells and utilised it to derive ES cells from non-obese diabetic (NOD) mice for the first time. This could dramatically reduce the number of animals used to study the genetic basis of type 1 diabetes and also has the potential to do the same for mouse models of other diseases.
ES cells are derived from early embryos and can be grown indefinitely in culture. They are a powerful tool in biomedical research because of their 'pluripotency', which means they can be transformed into all the cell types in the body, making them widely used in in vitro experiments and to generate disease models in mice.
Previously, understanding which genes play a role in type 1 diabetes involved breeding NOD mice with strains which had the gene of interest modified. This lengthy process required at least ten generations of breeding, involving many hundreds of animals, before the mice had a suitable genetic background for conducting the experiment.
The derivation of ES cells from the NOD mouse allows its genes to be directly manipulated to study type 1 diabetes without many generations of backcrossing, dramatically reducing the number of mice required per experiment. The NOD ES cells are now freely available to the research community, potentially reducing the number of mice used in type 1 diabetes research worldwide.
The new medium, which contains a novel mixture of cell growth factors and inhibitors but no animal products, has also allowed the derivation of ES cells from every strain of mouse tested so far with extremely high efficiency. Dr Nichols' laboratory has made this technology available so that it can be applied to other mouse models of disease where deriving ES cells has previously proven impossible.
Retinal explant tissue culture system
Prize winners: Dr Keith Martin and Mr Thomas Johnson, University of Cambridge
Dr Martin, and his colleague Mr Thomas Johnson, work investigates the potential of stem cells to protect vulnerable nerve cells in the injured retina. Their aim is to develop new treatments for glaucoma, the leading cause of irreversible blindness worldwide, and other eye diseases. Dr Martin and Mr Johnson pioneered a new method for retinal tissue culture that replaces the need for experiments on live animals.
They have shown that the cultured eye tissue remains healthy, maintains its layered architecture, and retains the ability to make new proteins. The tissue also responds to stem cell transplantation in a similar way to the eyes of living animals.
As well as replacing the use of live animals, the new method has brought about an eight-fold reduction in the number of animals used, because eight sections of tissue can be obtained from one rat.
Highly commended: Mr Charalambos Tymvios, Imperial College London
Mr Tymvious was commended for NC3Rs-funded research on refining a mouse model of pulmonary embolism.
Highly commended: Dr Jenny Morton, University of Cambridge
Dr Morton was highly commended for a paper describing the refinement of tests to measure cognitive deficits in mice used for neurodegenerative disease research.
DNA fingerprinting for Schistosoma parasites
Prize winner: Dr Charlotte Gower, Imperial College London
In her research, Dr Gower worked on schistosomes, the worm-like parasites that cause bilharzia, a tropical disease affecting an estimated 200 million people worldwide. The disease can be seriously debilitating, causing long term liver and intestinal damage, and can sometimes be fatal. Dr Gower was awarded the 3Rs Prize for a new application of DNA fingerprinting which replaces the need for using animals in this research area and improves the value and accuracy of her results.
Dr Gower has taken advantage of recent advances in how DNA can be stored at room temperature and characterised from minute samples in order to collect parasite DNA samples directly from infected people in areas where the disease is endemic. Previously, there was a need to grow the parasites in the laboratory for study by collecting worm eggs from human faeces and using them to infect snails and then rodents.
Highly Commended: Dr John Doe, Syngenta
Refining Leishmania infection in hamsters
Prize winners: Professor Alan Fairlamb and Dr Susan Wyllie, University of Dundee
In their research, Professor Fairlamb and Dr Wyllie infect hamsters with the parasite Leishmania donovani which causes visceral leishmaniasis. By using a different route of infection, the duration and severity of the disease in the hamster was reduced without compromising the quality of the scientific outcome.
Professor Fairlamb compared the commonly-used intracardial route of infection with the intraperitoneal route. The results showed that the intraperitoneal route is a simpler, safer and effective method of inoculating the hamsters.
Natural routes of bacterial infection in mice
Prize winner: Dr Siouxsie Wiles, Imperial College London
In her research, Dr Wiles infects mice with bacteria from the same family as E. coli to study the paths of infection. Traditionally, every mouse has been infected by putting a tube down its throat to deliver the bacteria to the stomach - a process called gavage.
Dr Wiles tried infecting only one mouse in this way, then putting it in a cage with uninfected mice and letting nature take its course.
The results showed higher infection rates than the traditional technique. But more importantly, the research was refined so that far fewer animals were subjected to gavage, and the new approach also reduced the total number of animals used by improving the reliability of infection.
Read more about our 3Rs Prize, including how to apply.